4KLV
 
 | | Crystal structure of a GNAT superfamily acetyltransferase PA4794 in complex with 4-methylumbelliferyl phosphate | | Descriptor: | 1,2-ETHANEDIOL, 4-methylumbelliferyl phosphate, SULFATE ION, ... | | Authors: | Majorek, K.A, Chruszcz, M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-05-07 | | Release date: | 2013-05-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural, Functional, and Inhibition Studies of a Gcn5-related N-Acetyltransferase (GNAT) Superfamily Protein PA4794: A NEW C-TERMINAL LYSINE PROTEIN ACETYLTRANSFERASE FROM PSEUDOMONAS AERUGINOSA.
J.Biol.Chem., 288, 2013
|
|
4KOT
 
 | | Crystal structure of a GNAT superfamily acetyltransferase PA4794 in complex with Cefotaxime | | Descriptor: | (6R,7R)-3-(acetyloxymethyl)-7-[[(2Z)-2-(2-amino-1,3-thiazol-4-yl)-2-methoxyimino-ethanoyl]amino]-8-oxo-5-thia-1-azabicy clo[4.2.0]oct-2-ene-2-carboxylic acid, 1,2-ETHANEDIOL, SULFATE ION, ... | | Authors: | Majorek, K.A, Chruszcz, M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-05-12 | | Release date: | 2013-06-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural, Functional, and Inhibition Studies of a Gcn5-related N-Acetyltransferase (GNAT) Superfamily Protein PA4794: A NEW C-TERMINAL LYSINE PROTEIN ACETYLTRANSFERASE FROM PSEUDOMONAS AERUGINOSA.
J.Biol.Chem., 288, 2013
|
|
4KOX
 
 | | Crystal structure of a GNAT superfamily acetyltransferase PA4794 in complex with Cefalotin | | Descriptor: | 1,2-ETHANEDIOL, CEPHALOTHIN, SULFATE ION, ... | | Authors: | Majorek, K.A, Chruszcz, M, Zimmerman, M.D, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-05-12 | | Release date: | 2013-06-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural, Functional, and Inhibition Studies of a Gcn5-related N-Acetyltransferase (GNAT) Superfamily Protein PA4794: A NEW C-TERMINAL LYSINE PROTEIN ACETYLTRANSFERASE FROM PSEUDOMONAS AERUGINOSA.
J.Biol.Chem., 288, 2013
|
|
4KOR
 
 | | Crystal structure of a GNAT superfamily acetyltransferase PA4794 in complex with 7-aminocephalosporanic acid | | Descriptor: | 1,2-ETHANEDIOL, 7-aminocephalosporanic acid, SULFATE ION, ... | | Authors: | Majorek, K.A, Porebski, P.J, Chruszcz, M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-05-12 | | Release date: | 2013-06-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural, Functional, and Inhibition Studies of a Gcn5-related N-Acetyltransferase (GNAT) Superfamily Protein PA4794: A NEW C-TERMINAL LYSINE PROTEIN ACETYLTRANSFERASE FROM PSEUDOMONAS AERUGINOSA.
J.Biol.Chem., 288, 2013
|
|
4KOV
 
 | | Crystal structure of a GNAT superfamily acetyltransferase PA4794 in complex with Cefuroxime | | Descriptor: | (6R,7R)-3-[(carbamoyloxy)methyl]-7-{[(2Z)-2-(furan-2-yl)-2-(methoxyimino)acetyl]amino}-8-oxo-5-thia-1-azabicyclo[4.2.0]oct-2-ene-2-carboxylic acid, 1,2-ETHANEDIOL, SULFATE ION, ... | | Authors: | Majorek, K.A, Chruszcz, M, Otwinowski, Z, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-05-12 | | Release date: | 2013-06-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural, Functional, and Inhibition Studies of a Gcn5-related N-Acetyltransferase (GNAT) Superfamily Protein PA4794: A NEW C-TERMINAL LYSINE PROTEIN ACETYLTRANSFERASE FROM PSEUDOMONAS AERUGINOSA.
J.Biol.Chem., 288, 2013
|
|
4KOU
 
 | | Crystal structure of a GNAT superfamily acetyltransferase PA4794 in complex with Cefixime | | Descriptor: | (6R,7R)-7-({(2Z)-2-(2-amino-1,3-thiazol-4-yl)-2-[(carboxymethoxy)imino]acetyl}amino)-3-ethenyl-8-oxo-5-thia-1-azabicyclo[4.2.0]oct-2-ene-2-carboxylic acid, 1,2-ETHANEDIOL, SULFATE ION, ... | | Authors: | Majorek, K.A, Chruszcz, M, Otwinowski, Z, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-05-12 | | Release date: | 2013-06-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural, Functional, and Inhibition Studies of a Gcn5-related N-Acetyltransferase (GNAT) Superfamily Protein PA4794: A NEW C-TERMINAL LYSINE PROTEIN ACETYLTRANSFERASE FROM PSEUDOMONAS AERUGINOSA.
J.Biol.Chem., 288, 2013
|
|
4L89
 
 | | Crystal structure of a GNAT superfamily acetyltransferase PA4794 in complex with covalently bound CoA | | Descriptor: | 1,2-ETHANEDIOL, COENZYME A, SULFATE ION, ... | | Authors: | Majorek, K.A, Chruszcz, M, Maclean, E, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-06-16 | | Release date: | 2013-07-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural, functional, and inhibition studies of a Gcn5-related N-acetyltransferase (GNAT) superfamily protein PA4794: a new C-terminal lysine protein acetyltransferase from pseudomonas aeruginosa.
J.Biol.Chem., 288, 2013
|
|
4KUB
 
 | | Crystal structure of a GNAT superfamily acetyltransferase PA4794 in complex with CoA | | Descriptor: | 1,2-ETHANEDIOL, COENZYME A, GNAT superfamily acetyltransferase PA4794, ... | | Authors: | Majorek, K.A, Chruszcz, M, Osinski, T, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-05-21 | | Release date: | 2013-06-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Structural, functional, and inhibition studies of a Gcn5-related N-acetyltransferase (GNAT) superfamily protein PA4794: a new C-terminal lysine protein acetyltransferase from pseudomonas aeruginosa.
J.Biol.Chem., 288, 2013
|
|
4KUA
 
 | | Crystal structure of a GNAT superfamily acetyltransferase PA4794 | | Descriptor: | 1,2-ETHANEDIOL, GNAT superfamily acetyltransferase PA4794, SULFATE ION | | Authors: | Majorek, K.A, Chruszcz, M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-05-21 | | Release date: | 2013-06-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural, functional, and inhibition studies of a Gcn5-related N-acetyltransferase (GNAT) superfamily protein PA4794: a new C-terminal lysine protein acetyltransferase from pseudomonas aeruginosa.
J.Biol.Chem., 288, 2013
|
|
4E4W
 
 | | Structure of the C-terminal domain of the Saccharomyces cerevisiae MUTL alpha (MLH1/PMS1) heterodimer | | Descriptor: | 1,2-ETHANEDIOL, DNA mismatch repair protein MLH1, DNA mismatch repair protein PMS1, ... | | Authors: | Gueneau, E, Legrand, P, Charbonnier, J.B. | | Deposit date: | 2012-03-13 | | Release date: | 2013-02-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the MutLalpha C-terminal domain reveals how Mlh1 contributes to Pms1 endonuclease site.
Nat.Struct.Mol.Biol., 20, 2013
|
|
6O2K
 
 | | Drosophila melanogaster CENP-C cupin domain | | Descriptor: | Centromeric protein-C, isoform A | | Authors: | Chik, J.K, Cho, U.S. | | Deposit date: | 2019-02-23 | | Release date: | 2019-08-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structures of CENP-C cupin domains at regional centromeres reveal unique patterns of dimerization and recruitment functions for the inner pocket.
J.Biol.Chem., 294, 2019
|
|
5YFS
 
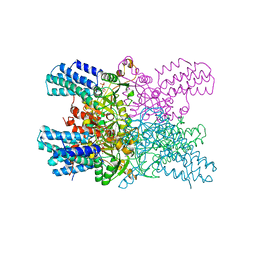 | | Crystal structure of ribose-1,5-bisphosphate isomerase mutant C135S from Pyrococcus horikoshii OT3 in complex with ribose-1,5-bisphosphate | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,5-di-O-phosphono-alpha-D-ribofuranose, CHLORIDE ION, ... | | Authors: | Gogoi, P, Kanaujia, S.P. | | Deposit date: | 2017-09-22 | | Release date: | 2018-02-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A presumed homologue of the regulatory subunits of eIF2B functions as ribose-1,5-bisphosphate isomerase in Pyrococcus horikoshii OT3.
Sci Rep, 8, 2018
|
|
6E4T
 
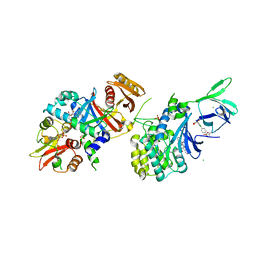 | | Structure of AMPK bound to activator | | Descriptor: | 1-O-{6-chloro-5-[4-(1-hydroxycyclobutyl)phenyl]-1H-indole-3-carbonyl}-beta-D-glucopyranuronic acid, 5'-AMP-activated protein kinase catalytic subunit alpha-1, 5'-AMP-activated protein kinase subunit beta-1, ... | | Authors: | Calabrese, M.F, Kurumbail, R.G. | | Deposit date: | 2018-07-18 | | Release date: | 2018-08-08 | | Last modified: | 2018-09-05 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Acyl Glucuronide Metabolites of 6-Chloro-5-[4-(1-hydroxycyclobutyl)phenyl]-1 H-indole-3-carboxylic Acid (PF-06409577) and Related Indole-3-carboxylic Acid Derivatives are Direct Activators of Adenosine Monophosphate-Activated Protein Kinase (AMPK).
J. Med. Chem., 61, 2018
|
|
6E4U
 
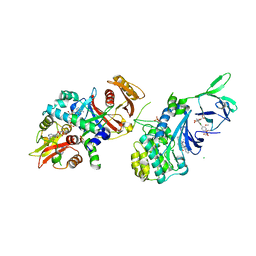 | | Structure of AMPK bound to activator | | Descriptor: | 1-O-{6-chloro-5-[6-(dimethylamino)-2-methoxypyridin-3-yl]-1H-indole-3-carbonyl}-beta-D-glucopyranuronic acid, 5'-AMP-activated protein kinase catalytic subunit alpha-1, 5'-AMP-activated protein kinase subunit beta-1, ... | | Authors: | Calabrese, M.F, Kurumbail, R.G. | | Deposit date: | 2018-07-18 | | Release date: | 2018-08-08 | | Last modified: | 2018-09-05 | | Method: | X-RAY DIFFRACTION (3.27 Å) | | Cite: | Acyl Glucuronide Metabolites of 6-Chloro-5-[4-(1-hydroxycyclobutyl)phenyl]-1 H-indole-3-carboxylic Acid (PF-06409577) and Related Indole-3-carboxylic Acid Derivatives are Direct Activators of Adenosine Monophosphate-Activated Protein Kinase (AMPK).
J. Med. Chem., 61, 2018
|
|
2PA8
 
 | |
6O58
 
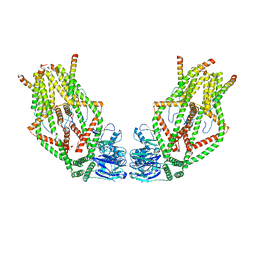 | | Human MCU-EMRE complex, dimer of channel | | Descriptor: | CALCIUM ION, Calcium uniporter protein, mitochondrial, ... | | Authors: | Wang, Y, Bai, X, Jiang, Y. | | Deposit date: | 2019-03-01 | | Release date: | 2019-05-22 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural Mechanism of EMRE-Dependent Gating of the Human Mitochondrial Calcium Uniporter.
Cell, 177, 2019
|
|
5YG7
 
 | | Crystal structure of ribose-1,5-bisphosphate isomerase mutant D204N from Pyrococcus horikoshii OT3 in complex with ribose-1,5-bisphosphate and GMP | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 1,5-di-O-phosphono-alpha-D-ribofuranose, GUANOSINE-5'-MONOPHOSPHATE, ... | | Authors: | Gogoi, P, Kanaujia, S.P. | | Deposit date: | 2017-09-22 | | Release date: | 2018-02-14 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A presumed homologue of the regulatory subunits of eIF2B functions as ribose-1,5-bisphosphate isomerase in Pyrococcus horikoshii OT3.
Sci Rep, 8, 2018
|
|
6OFE
 
 | |
6OOH
 
 | | CTX-M-27 Beta Lactamase with Compound 14 | | Descriptor: | 3-(1H-pyrazol-1-yl)-N-[3-(1H-tetrazol-5-yl)phenyl]-5-(trifluoromethyl)benzamide, Beta-lactamase | | Authors: | Kemp, M, Chen, Y. | | Deposit date: | 2019-04-23 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.499 Å) | | Cite: | An Empirical Study of Amide-Heteroarene pi-Stacking Interactions Using Reversible Inhibitors of a Bacterial Serine Hydrolase.
Org Chem Front, 6, 2019
|
|
8CRO
 
 | | Cryo-EM structure of Pyrococcus furiosus transcription elongation complex | | Descriptor: | DNA Non-Template Strand, DNA Template Strand, DNA-directed RNA polymerase subunit Rpo10, ... | | Authors: | Tarau, D.M, Grunberger, F, Reichelt, R, Heiss, F.B, Pilsl, M, Hausner, W, Engel, C, Grohmann, D. | | Deposit date: | 2023-03-08 | | Release date: | 2024-04-17 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of archaeal RNA polymerase transcription elongation and Spt4/5 recruitment.
Nucleic Acids Res., 52, 2024
|
|
8CQW
 
 | | Crystal structure of the Candida albicans 80S ribosome in complex with Hygromycin B | | Descriptor: | 18S, 25S, 3-O-acetyl-2-O-(3-O-acetyl-6-deoxy-beta-D-glucopyranosyl)-6-deoxy-1-O-{[(2R,2'S,3a'R,4''S,5''R,6'S,7a'S)-5''-methyl-4''-{[(2E)-3-phenylprop-2-enoyl]oxy}decahydrodispiro[oxirane-2,3'-[1]benzofuran-2',2''-pyran]-6'-yl]carbonyl}-beta-D-glucopyranose, ... | | Authors: | Kolosova, O, Zgadzay, Y, Yusupov, M. | | Deposit date: | 2023-03-07 | | Release date: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Drug-induced rotational movement of the ribosome is a key factor for read-through enhancement
To Be Published
|
|
8CRE
 
 | | Crystal structure of the Candida albicans 80S ribosome in complex with geneticin G418 | | Descriptor: | 18S, 25S, 3-O-acetyl-2-O-(3-O-acetyl-6-deoxy-beta-D-glucopyranosyl)-6-deoxy-1-O-{[(2R,2'S,3a'R,4''S,5''R,6'S,7a'S)-5''-methyl-4''-{[(2E)-3-phenylprop-2-enoyl]oxy}decahydrodispiro[oxirane-2,3'-[1]benzofuran-2',2''-pyran]-6'-yl]carbonyl}-beta-D-glucopyranose, ... | | Authors: | Kolosova, O, Zgadzay, Y, Yusupov, M. | | Deposit date: | 2023-03-08 | | Release date: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Drug-induced rotational movement of the ribosome is a key factor for read-through enhancement
To Be Published
|
|
5YFX
 
 | | Crystal structure of ribose-1,5-bisphosphate isomerase mutant D204N from Pyrococcus horikoshii OT3 in complex with ribose-1,5-bisphosphate and AMP | | Descriptor: | 1,5-di-O-phosphono-alpha-D-ribofuranose, ADENOSINE MONOPHOSPHATE, Ribose 1,5-bisphosphate isomerase | | Authors: | Gogoi, P, Kanaujia, S.P. | | Deposit date: | 2017-09-22 | | Release date: | 2018-02-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A presumed homologue of the regulatory subunits of eIF2B functions as ribose-1,5-bisphosphate isomerase in Pyrococcus horikoshii OT3.
Sci Rep, 8, 2018
|
|
6E8M
 
 | |
6OHC
 
 | | E. coli Guanine Deaminase | | Descriptor: | GLYCEROL, Guanine deaminase, ZINC ION | | Authors: | Shek, R.S, French, J.B. | | Deposit date: | 2019-04-05 | | Release date: | 2019-07-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Determinants for Substrate Selectivity in Guanine Deaminase Enzymes of the Amidohydrolase Superfamily.
Biochemistry, 58, 2019
|
|
