4W4W
 
 | | JNK2/3 in complex with N-(2-methylpyridin-4-yl)-3-{4-[(phenylcarbamoyl)amino]-1H-pyrazol-1-yl}benzamide | | Descriptor: | N-(2-methylpyridin-4-yl)-3-{4-[(phenylcarbamoyl)amino]-1H-pyrazol-1-yl}benzamide, c-Jun N-terminal kinase 3 | | Authors: | Park, H, Iqbal, S, Hernandez, P, Mora, R, Zheng, K, Feng, Y, LoGrasso, P. | | Deposit date: | 2014-08-15 | | Release date: | 2015-02-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis and Biological Consequences for JNK2/3 Isoform Selective Aminopyrazoles.
Sci Rep, 5, 2015
|
|
4WHZ
 
 | |
1OWR
 
 | |
5AWM
 
 | |
1PZU
 
 | | An asymmetric NFAT1-RHR homodimer on a pseudo-palindromic, Kappa-B site | | Descriptor: | 5'-D(*AP*AP*TP*GP*GP*AP*AP*AP*TP*TP*CP*CP*TP*C)-3', 5'-D(*TP*TP*GP*AP*GP*GP*AP*AP*TP*TP*TP*CP*CP*A)-3', Nuclear factor of activated T-cells, ... | | Authors: | Jin, L, Sliz, P, Chen, L, Macian, F, Rao, A, Hogan, P.G, Harrison, S.C. | | Deposit date: | 2003-07-14 | | Release date: | 2003-09-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | An asymmetric NFAT1 dimer on a pseudo-palindromic KB-like DNA site
Nat.Struct.Biol., 10, 2003
|
|
4Z9L
 
 | | THE STRUCTURE OF JNK3 IN COMPLEX WITH AN IMIDAZOLE-PYRIMIDINE INHIBITOR | | Descriptor: | CYCLOHEXYL-{4-[5-(3,4-DICHLOROPHENYL)-2-PIPERIDIN-4-YL-3-PROPYL-3H-IMIDAZOL-4-YL]-PYRIMIDIN-2-YL}AMINE, Mitogen-activated protein kinase 10, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Scapin, G, Patel, S.B, Lisnock, J, Becker, J.W, Lograsso, P.V, Smart, O.S, Bricogne, G. | | Deposit date: | 2015-04-10 | | Release date: | 2015-05-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structure of JNK3 in complex with small molecule inhibitors: structural basis for potency and selectivity.
Chem.Biol., 10, 2003
|
|
2W83
 
 | | Crystal structure of the ARF6 GTPase in complex with a specific effector, JIP4 | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, ADP-RIBOSYLATION FACTOR 6, C-JUN-AMINO-TERMINAL KINASE-INTERACTING PROTEIN 4, ... | | Authors: | Isabet, T, Montagnac, G, Regazzoni, K, Raynal, B, El Khadali, F, Franco, M, England, P, Chavrier, P, Houdusse, A, Menetrey, J. | | Deposit date: | 2009-01-08 | | Release date: | 2009-07-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | The Structural Basis of Arf Effector Specificity: The Crystal Structure of Arf6 in a Complex with Jip4.
Embo J., 28, 2009
|
|
6UCI
 
 | | Transcription factor DeltaFosB bZIP domain self-assembly, oxidized form | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Protein fosB | | Authors: | Yin, Z, Machius, M, Rudenko, G. | | Deposit date: | 2019-09-16 | | Release date: | 2020-01-15 | | Last modified: | 2020-07-01 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Self-assembly of the bZIP transcription factor Delta FosB.
Curr Res Struct Biol, 2, 2020
|
|
6R7J
 
 | | Ligand complex of RORg LBD | | Descriptor: | (2~{R})-2-acetamido-2-(4-ethylsulfonylphenyl)-~{N}-[4-[1,1,1,3,3,3-hexakis(fluoranyl)-2-oxidanyl-propan-2-yl]phenyl]ethanamide, DIMETHYL SULFOXIDE, Nuclear receptor ROR-gamma, ... | | Authors: | Xue, Y, Aagaard, A, Narjes, F. | | Deposit date: | 2019-03-29 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Discovery of Potent and Orally Bioavailable Inverse Agonists of the Retinoic Acid Receptor-Related Orphan Receptor C2.
Acs Med.Chem.Lett., 10, 2019
|
|
7YMD
 
 | | Cryo-EM structure of Nse1/3/4 | | Descriptor: | Non-structural maintenance of chromosome element 3, Non-structural maintenance of chromosome element 4, Non-structural maintenance of chromosomes element 1 | | Authors: | Qian, L, Jun, Z, Zhenguo, C, Wang, L. | | Deposit date: | 2022-07-28 | | Release date: | 2024-01-31 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (4.176 Å) | | Cite: | Cryo-EM structures of Smc5/6 in multiple states reveal its assembly and functional mechanisms.
Nat.Struct.Mol.Biol., 2024
|
|
7YQH
 
 | | Cryo-EM structure of 8-subunit Smc5/6 | | Descriptor: | DNA repair protein KRE29, E3 SUMO-protein ligase MMS21, Non-structural maintenance of chromosome element 3, ... | | Authors: | Qian, L, Jun, Z, Xiang, Z, Tong, C, Zhaoning, W, Duo, J, Zhenguo, C, Lanfeng, W. | | Deposit date: | 2022-08-07 | | Release date: | 2024-01-31 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | Cryo-EM structures of Smc5/6 in multiple states reveal its assembly and functional mechanisms.
Nat.Struct.Mol.Biol., 2024
|
|
5Z1D
 
 | | MAP2K7 C276S mutant-inhibitor | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Dual specificity mitogen-activated protein kinase kinase 7, N-[3-(6-methyl-1H-indazol-3-yl)phenyl]prop-2-enamide | | Authors: | Kinoshita, T, London, N. | | Deposit date: | 2017-12-26 | | Release date: | 2019-01-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Covalent Docking Identifies a Potent and Selective MKK7 Inhibitor.
Cell Chem Biol, 26, 2019
|
|
5Z1E
 
 | | MAP2K7 C218S mutant-inhibitor | | Descriptor: | Dual specificity mitogen-activated protein kinase kinase 7, N-[3-(6-methyl-1H-indazol-3-yl)phenyl]prop-2-enamide | | Authors: | Kinoshita, T, London, N. | | Deposit date: | 2017-12-26 | | Release date: | 2019-01-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Covalent Docking Identifies a Potent and Selective MKK7 Inhibitor.
Cell Chem Biol, 26, 2019
|
|
1PMN
 
 | | Crystal structure of JNK3 in complex with an imidazole-pyrimidine inhibitor | | Descriptor: | CYCLOPROPYL-{4-[5-(3,4-DICHLOROPHENYL)-2-[(1-METHYL)-PIPERIDIN]-4-YL-3-PROPYL-3H-IMIDAZOL-4-YL]-PYRIMIDIN-2-YL}AMINE, Mitogen-activated protein kinase 10 | | Authors: | Scapin, G, Patel, S.B, Lisnock, J, Becker, J.W, LoGrasso, P.V. | | Deposit date: | 2003-06-11 | | Release date: | 2003-09-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of JNK3 in complex with small molecule inhibitors: structural basis for potency and selectivity
Chem.Biol., 10, 2003
|
|
1PMV
 
 | | The structure of JNK3 in complex with a dihydroanthrapyrazole inhibitor | | Descriptor: | 2,6-DIHYDROANTHRA/1,9-CD/PYRAZOL-6-ONE, Mitogen-activated protein kinase 10 | | Authors: | Scapin, G, Patel, S.B, Lisnock, J, Becker, J.W, LoGrasso, P.V. | | Deposit date: | 2003-06-11 | | Release date: | 2003-09-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structure of JNK3 in complex with small molecule inhibitors: structural basis for potency and selectivity
Chem.Biol., 10, 2003
|
|
1PMU
 
 | | The crystal structure of JNK3 in complex with a phenantroline inhibitor | | Descriptor: | 9-(4-HYDROXYPHENYL)-2,7-PHENANTHROLINE, CHLORIDE ION, Mitogen-activated protein kinase 10 | | Authors: | Scapin, G, Patel, S.B, Lisnock, J, Becker, J.W, LoGrasso, P.V. | | Deposit date: | 2003-06-11 | | Release date: | 2003-09-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The structure of JNK3 in complex with small molecule inhibitors: structural basis for potency and selectivity
Chem.Biol., 10, 2003
|
|
7CBX
 
 | |
5B2L
 
 | | A crucial role of Cys218 in the stabilization of an unprecedented auto-inhibition form of MAP2K7 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Dual specificity mitogen-activated protein kinase kinase 7, GLYCEROL | | Authors: | Sogabe, Y, Hashimoto, T, Matsumoto, T, Kirii, Y, Sawa, M, Kinoshita, T. | | Deposit date: | 2016-01-19 | | Release date: | 2016-04-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A crucial role of Cys218 in configuring an unprecedented auto-inhibition form of MAP2K7
Biochem.Biophys.Res.Commun., 473, 2016
|
|
5B2K
 
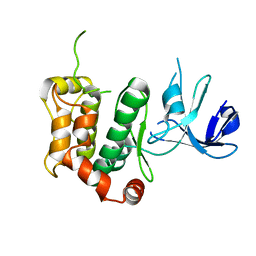 | | A crucial role of Cys218 in the stabilization of an unprecedented auto-inhibition form of MAP2K7 | | Descriptor: | Dual specificity mitogen-activated protein kinase kinase 7 | | Authors: | Sogabe, Y, Hashimoto, T, Matsumoto, T, Kirii, Y, Sawa, M, Kinoshita, T. | | Deposit date: | 2016-01-19 | | Release date: | 2016-04-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | A crucial role of Cys218 in configuring an unprecedented auto-inhibition form of MAP2K7
Biochem.Biophys.Res.Commun., 473, 2016
|
|
5B2M
 
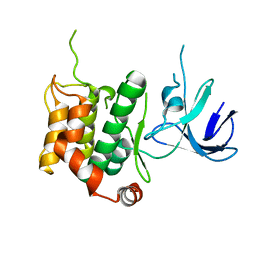 | | A crucial role of Cys218 in the stabilization of an unprecedented auto-inhibition form of MAP2K7 | | Descriptor: | Dual specificity mitogen-activated protein kinase kinase 7 | | Authors: | Sogabe, Y, Hashimoto, T, Matsumoto, T, Kirii, Y, Sawa, M, Kinoshita, T. | | Deposit date: | 2016-01-19 | | Release date: | 2016-04-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | A crucial role of Cys218 in configuring an unprecedented auto-inhibition form of MAP2K7
Biochem.Biophys.Res.Commun., 473, 2016
|
|
4PXJ
 
 | |
8PR4
 
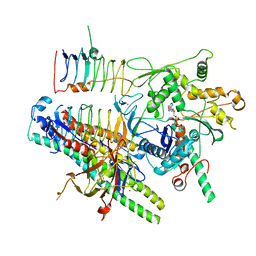 | | Dynactin pointed end bound to JIP3 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Arp11, C-Jun-amino-terminal kinase-interacting protein 3, ... | | Authors: | Singh, K, Lau, C.K, Manigrasso, G, Gassmann, R, Carter, A.P. | | Deposit date: | 2023-07-12 | | Release date: | 2024-03-27 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Molecular mechanism of dynein-dynactin complex assembly by LIS1.
Science, 383, 2024
|
|
8PTK
 
 | | Composite structure of Dynein-Dynactin-JIP3-LIS1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ARP1 actin related protein 1 homolog A, ... | | Authors: | Singh, K, Lau, C.K, Manigrasso, G, Gassmann, R, Carter, A.P. | | Deposit date: | 2023-07-14 | | Release date: | 2024-03-27 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (10 Å) | | Cite: | Molecular mechanism of dynein-dynactin complex assembly by LIS1.
Science, 383, 2024
|
|
6L6Z
 
 | |
6FUZ
 
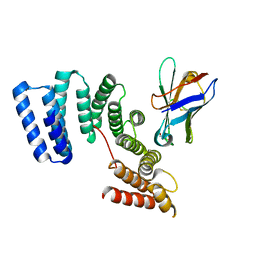 | | Crystal structure of the TPR domain of KLC1 in complex with the C-terminal peptide of JIP1 | | Descriptor: | GLYCEROL, Kinesin light chain 1,Kinesin light chain 1,C-Jun-amino-terminal kinase-interacting protein 1, nanobody | | Authors: | Pernigo, S, Dodding, M.P, Steiner, R.A. | | Deposit date: | 2018-02-28 | | Release date: | 2018-05-02 | | Last modified: | 2019-09-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for isoform-specific kinesin-1 recognition of Y-acidic cargo adaptors.
Elife, 7, 2018
|
|
