6EJQ
 
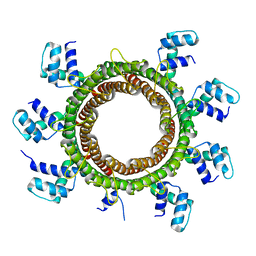 | |
6EO7
 
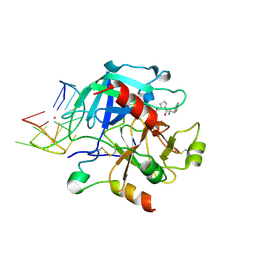 | | X-ray structure of the complex between human alpha-thrombin and modified 15-mer DNA aptamer containing 5-(3-(acetamide-N-yl)-1-propen-1-yl)-2'-deoxyuridine residue | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, GA68B2 - MODIFIED HUMAN THROMBIN BINDING APTAMER, ... | | Authors: | Dolot, R.M, Nawrot, B, Yang, X. | | Deposit date: | 2017-10-09 | | Release date: | 2017-10-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Crystal structures of thrombin in complex with chemically modified thrombin DNA aptamers reveal the origins of enhanced affinity.
Nucleic Acids Res., 46, 2018
|
|
6EQL
 
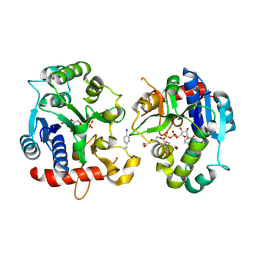 | | Crystal Structure of Human Glycogenin-1 (GYG1) Tyr195pIPhe mutant complexed with manganese and UDP | | Descriptor: | 1,2-ETHANEDIOL, Glycogenin-1, MANGANESE (II) ION, ... | | Authors: | Bailey, H.J, Kopec, J, Bilyard, M.K, Bezerra, G.A, Seo Lee, S, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Davis, B.G, Yue, W.W. | | Deposit date: | 2017-10-13 | | Release date: | 2017-12-20 | | Last modified: | 2018-11-14 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Palladium-mediated enzyme activation suggests multiphase initiation of glycogenesis.
Nature, 563, 2018
|
|
6EQO
 
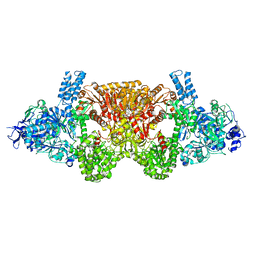 | | Tri-functional propionyl-CoA synthase of Erythrobacter sp. NAP1 with bound NADP+ and phosphomethylphosphonic acid adenylate ester | | Descriptor: | Acetyl-coenzyme A synthetase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER | | Authors: | Zarzycki, J, Bernhardsgruetter, I, Voegeli, B, Wagner, T, Engilberge, S, Girard, E, Shima, S, Erb, T.J. | | Deposit date: | 2017-10-13 | | Release date: | 2018-10-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The multicatalytic compartment of propionyl-CoA synthase sequesters a toxic metabolite.
Nat. Chem. Biol., 14, 2018
|
|
6KER
 
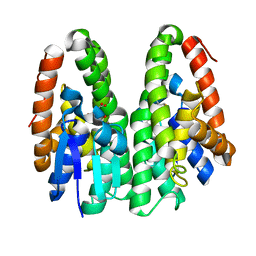 | | Crystal structure of D113A mutant of Drosophila melanogaster Noppera-bo, glutathione S-transferase epsilon 14 (DmGSTE14), in glutathione-bound form | | Descriptor: | GLUTATHIONE, Glutathione S-transferase E14 | | Authors: | Koiwai, K, Inaba, K, Morohashi, K, Yumoto, F, Niwa, R, Senda, T. | | Deposit date: | 2019-07-04 | | Release date: | 2019-10-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | An integrated approach to unravel a crucial structural property required for the function of the insect steroidogenic Halloween protein Noppera-bo.
J.Biol.Chem., 295, 2020
|
|
6K0S
 
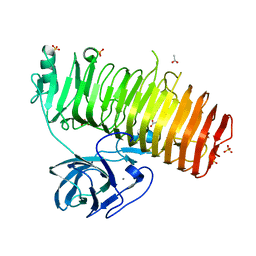 | | Catalytic domain of GH87 alpha-1,3-glucanase D1069A in complex with nigerose | | Descriptor: | ACETIC ACID, Alpha-1,3-glucanase, CALCIUM ION, ... | | Authors: | Itoh, T, Intuy, R, Suyotha, W, Hayashi, J, Yano, S, Makabe, K, Wakayama, M, Hibi, T. | | Deposit date: | 2019-05-07 | | Release date: | 2019-12-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.534 Å) | | Cite: | Structural insights into substrate recognition and catalysis by glycoside hydrolase family 87 alpha-1,3-glucanase from Paenibacillus glycanilyticus FH11.
Febs J., 287, 2020
|
|
6K16
 
 | |
6K31
 
 | |
6KK3
 
 | | Crystal structure of Zika NS2B-NS3 protease with compound 4 | | Descriptor: | 1-[(10~{R},17~{S},20~{S})-17,20-bis(4-azanylbutyl)-4,9,16,19,22-pentakis(oxidanylidene)-3,8,15,18,21-pentazabicyclo[22.2.2]octacosa-1(26),24,27-trien-10-yl]guanidine, Genome polyprotein | | Authors: | Quek, J.P. | | Deposit date: | 2019-07-23 | | Release date: | 2020-06-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure-Based Macrocyclization of Substrate Analogue NS2B-NS3 Protease Inhibitors of Zika, West Nile and Dengue viruses.
Chemmedchem, 15, 2020
|
|
6KKV
 
 | |
6K7R
 
 | | Crystal structure of thymidylate synthase from shrimp | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, Thymidylate synthase | | Authors: | Ma, Q, Zang, K, Liu, C. | | Deposit date: | 2019-06-08 | | Release date: | 2020-06-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structural analysis of a shrimp thymidylate synthase reveals species-specific interactions with dUMP and raltitrexed.
J Oceanol Limnol, 38, 2020
|
|
6JW9
 
 | |
6KAV
 
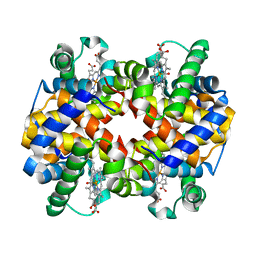 | | Carbonmonoxy human hemoglobin A in the R2 quaternary structure at 140 K: Light | | Descriptor: | CARBON MONOXIDE, Hemoglobin subunit alpha, Hemoglobin subunit beta, ... | | Authors: | Shibayama, N, Park, S.Y, Ohki, M, Sato-Tomita, A. | | Deposit date: | 2019-06-24 | | Release date: | 2020-02-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Direct observation of ligand migration within human hemoglobin at work.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6KB2
 
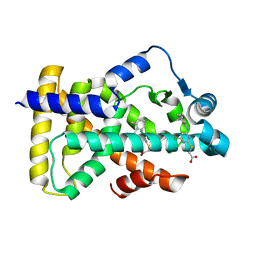 | | X-ray structure of human PPARalpha ligand binding domain-Wy14643 co-crystals obtained by soaking | | Descriptor: | 2-({4-CHLORO-6-[(2,3-DIMETHYLPHENYL)AMINO]PYRIMIDIN-2-YL}SULFANYL)ACETIC ACID, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Suda, K, Saito, K, Oyama, T, Ishii, I. | | Deposit date: | 2019-06-24 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
6KB9
 
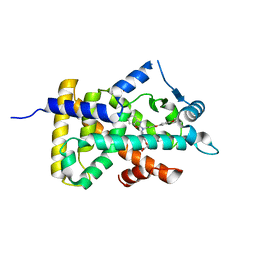 | | X-ray structure of human PPARalpha ligand binding domain-pemafibrate co-crystals obtained by cross-seeding | | Descriptor: | (2~{R})-2-[3-[[1,3-benzoxazol-2-yl-[3-(4-methoxyphenoxy)propyl]amino]methyl]phenoxy]butanoic acid, GLYCEROL, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Saito, K, Honda, A, Ishikawa, R, Oyama, T, Ishii, I. | | Deposit date: | 2019-06-24 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
6KG2
 
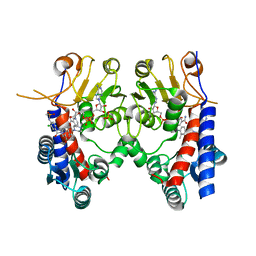 | | Human MTHFD2 in complex with Compound 18 | | Descriptor: | Bifunctional methylenetetrahydrofolate dehydrogenase/cyclohydrolase, mitochondrial, N-[2-chloranyl-4-[[7-methyl-8-(4-methylpiperazin-1-yl)-5-oxidanylidene-2,4-dihydro-1H-chromeno[3,4-c]pyridin-3-yl]carbonyl]phenyl]methanesulfonamide, ... | | Authors: | Suzuki, M, Matsui, Y, Ota, M, Kawai, J. | | Deposit date: | 2019-07-10 | | Release date: | 2019-11-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Discovery of a Potent, Selective, and Orally Available MTHFD2 Inhibitor (DS18561882) with in Vivo Antitumor Activity.
J.Med.Chem., 62, 2019
|
|
6KGH
 
 | | Crystal structure of Penicillin binding protein 3 (PBP3) from Mycobacterium tuerculosis (apo-form) | | Descriptor: | COBALT (II) ION, Penicillin-binding protein PbpB, SODIUM ION | | Authors: | Lu, Z.K, Zhang, A.L, Liu, X, Guddat, L, Yang, H.T, Rao, Z.H. | | Deposit date: | 2019-07-11 | | Release date: | 2020-03-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.108 Å) | | Cite: | Structures ofMycobacterium tuberculosisPenicillin-Binding Protein 3 in Complex with Fivebeta-Lactam Antibiotics Reveal Mechanism of Inactivation.
Mol.Pharmacol., 97, 2020
|
|
5MPV
 
 | |
5MPZ
 
 | |
6R9R
 
 | | Crystal structure of Csx1 in complex with cyclic oligoadenylate cOA4 conformation 2 | | Descriptor: | CRISPR-associated (Cas) DxTHG family, circular RNA (5'-R(P*AP*AP*AP*A)-3') | | Authors: | Molina, R, Montoya, G, Sofos, N, Stella, S. | | Deposit date: | 2019-04-03 | | Release date: | 2019-10-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of Csx1-cOA4complex reveals the basis of RNA decay in Type III-B CRISPR-Cas.
Nat Commun, 10, 2019
|
|
5MQ4
 
 | | Crystal Structure of the leucine zipper of human PRKCBP1 | | Descriptor: | Protein kinase C-binding protein 1, SULFATE ION, ZINC ION | | Authors: | Krojer, T, Savitsky, P, Picaud, S, Newman, J, Tallant, C, Heroven, C, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Filippakopoulos, P. | | Deposit date: | 2016-12-20 | | Release date: | 2017-01-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of the leucine zipper of human PRKCBP1
To Be Published
|
|
6RAS
 
 | | Pmar-Lig_Pre. | | Descriptor: | ADENOSINE MONOPHOSPHATE, ATP-dependent DNA ligase, DNA, ... | | Authors: | Leiros, H.K.S, Williamson, A. | | Deposit date: | 2019-04-07 | | Release date: | 2019-07-10 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural intermediates of a DNA-ligase complex illuminate the role of the catalytic metal ion and mechanism of phosphodiester bond formation.
Nucleic Acids Res., 47, 2019
|
|
5MSY
 
 | | Glycoside hydrolase BT_1012 | | Descriptor: | AMMONIA, Glycoside hydrolase, PHOSPHATE ION | | Authors: | Basle, A, Ndeh, D, Rogowski, A, Cartmell, A, Luis, A.S, Venditto, I, Labourel, A, Gilbert, H.J. | | Deposit date: | 2017-01-06 | | Release date: | 2017-03-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Complex pectin metabolism by gut bacteria reveals novel catalytic functions.
Nature, 544, 2017
|
|
5MQO
 
 | | Glycoside hydrolase BT_1003 | | Descriptor: | Non-reducing end beta-L-arabinofuranosidase | | Authors: | Basle, A, Ndeh, D, Rogowski, A, Cartmell, A, Luis, A.S, Venditto, I, Labourel, A, Gilbert, H.J. | | Deposit date: | 2016-12-20 | | Release date: | 2017-03-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Complex pectin metabolism by gut bacteria reveals novel catalytic functions.
Nature, 544, 2017
|
|
5M9A
 
 | | Human angiogenin PD variant H13R | | Descriptor: | Angiogenin, D(-)-TARTARIC ACID | | Authors: | Bradshaw, W.J, Rehman, S, Pham, T.T.K, Thiyagarajan, N, Lee, R.L, Subramanian, V, Acharya, K.R. | | Deposit date: | 2016-11-01 | | Release date: | 2017-02-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural insights into human angiogenin variants implicated in Parkinson's disease and Amyotrophic Lateral Sclerosis.
Sci Rep, 7, 2017
|
|
