8WKJ
 
 | |
7QJL
 
 | | Crystal structure of CYP142 from Mycobacterium tuberculosis in complex with an inhibitor | | Descriptor: | ACETATE ION, BROMIDE ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Snee, M, Levy, C, Katariya, M. | | Deposit date: | 2021-12-16 | | Release date: | 2022-12-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Structure Based Discovery of Inhibitors of CYP125 and CYP142 from Mycobacterium tuberculosis.
Chemistry, 29, 2023
|
|
2WRT
 
 | | The 2.4 Angstrom structure of the Fasciola hepatica mu class GST, GST26 | | Descriptor: | CHLORIDE ION, GLUTATHIONE S-TRANSFERASE CLASS-MU 26 KDA ISOZYME 51 | | Authors: | Line, K, Isupov, M.N, LaCourse, E.J, Brophy, P.M, Littlechild, J.A. | | Deposit date: | 2009-09-02 | | Release date: | 2010-09-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The 2.5 Angstrom Structure of a Mu Class Gst from Fasciola Hepatica
To be Published
|
|
5ET6
 
 | | Human muscle fructose-1,6-bisphosphatase in inactive T-state in complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Fructose-1,6-bisphosphatase isozyme 2 | | Authors: | Barciszewski, J, Wisniewski, J, Kolodziejczyk, R, Dzugaj, A, Jaskolski, M, Rakus, D. | | Deposit date: | 2015-11-17 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.845 Å) | | Cite: | T-to-R switch of muscle fructose-1,6-bisphosphatase involves fundamental changes of secondary and quaternary structure.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
3E6L
 
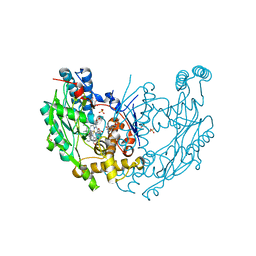 | | Structure of murine INOS oxygenase domain with inhibitor AR-C132283 | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ETHYL 4-[(4-CHLOROPYRIDIN-2-YL)AMINO]PIPERIDINE-1-CARBOXYLATE, Nitric oxide synthase, ... | | Authors: | Garcin, E.D, Arvai, A.S, Rosenfeld, R.J, Kroeger, M.D, Crane, B.R, Andersson, G, Andrews, G, Hamley, P.J, Mallinder, P.R, Nicholls, D.J, St-Gallay, S.A, Tinker, A.C, Gensmantel, N.P, Mete, A, Cheshire, D.R, Connolly, S, Stueh, D.J, Aberg, A, Wallace, A.V, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2008-08-15 | | Release date: | 2008-10-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Anchored plasticity opens doors for selective inhibitor design in nitric oxide synthase.
Nat.Chem.Biol., 4, 2008
|
|
7JQ0
 
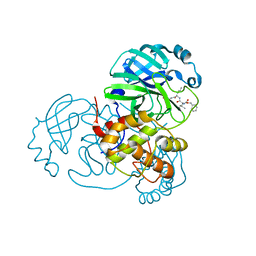 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI3 | | Descriptor: | 3C-like proteinase, N-[(benzyloxy)carbonyl]-L-valyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide | | Authors: | Yang, K, Liu, W. | | Deposit date: | 2020-08-10 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | A Quick Route to Multiple Highly Potent SARS-CoV-2 Main Protease Inhibitors*.
Chemmedchem, 16, 2021
|
|
5ET7
 
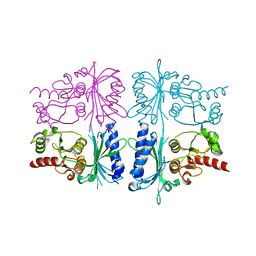 | | Human muscle fructose-1,6-bisphosphatase in inactive T-state | | Descriptor: | Fructose-1,6-bisphosphatase isozyme 2 | | Authors: | Barciszewski, J, Wisniewski, J, Kolodziejczyk, R, Dzugaj, A, Jaskolski, M, Rakus, D. | | Deposit date: | 2015-11-17 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.989 Å) | | Cite: | T-to-R switch of muscle fructose-1,6-bisphosphatase involves fundamental changes of secondary and quaternary structure.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
2WJL
 
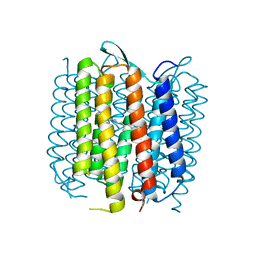 | | Bacteriorhodopsin mutant E194D | | Descriptor: | Bacteriorhodopsin, RETINAL | | Authors: | Potschies, M, Wolf, S, Freier, E, Hofmann, E, Gerwert, K. | | Deposit date: | 2009-05-27 | | Release date: | 2010-08-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Directional proton transfer in membrane proteins achieved through protonated protein-bound water molecules: a proton diode.
Angew. Chem. Int. Ed. Engl., 49, 2010
|
|
3E74
 
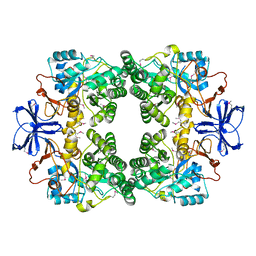 | |
4N2Q
 
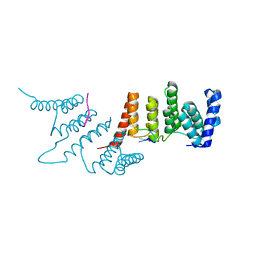 | | Crystal structure of THA8 in complex with Zm4 RNA | | Descriptor: | RNA (5'-R(*AP*AP*GP*AP*AP*GP*AP*AP*AP*UP*UP*GP*G)-3'), THA8 RNA binding protein | | Authors: | Ke, J, Chen, R.Z, Ban, T, Zhou, X.E, Gu, X, Brunzelle, J.S, Zhu, J.K, Melcher, K, Xu, H.E. | | Deposit date: | 2013-10-06 | | Release date: | 2013-10-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for RNA recognition by a dimeric PPR-protein complex.
Nat.Struct.Mol.Biol., 20, 2013
|
|
4MTB
 
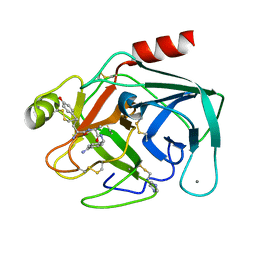 | |
8WOU
 
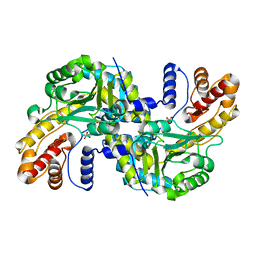 | |
2WUC
 
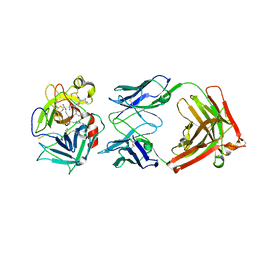 | | Crystal structure of HGFA in complex with the allosteric non- inhibitory antibody Fab40.deltaTrp and Ac-KQLR-chloromethylketone | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACE-KQLR-CHLOROMETHYLKETONE INHIBITOR, FAB FRAGMENT FAB40.DELTATRP HEAVY CHAIN, ... | | Authors: | Ganesan, R, Eigenbrot, C, Shia, S. | | Deposit date: | 2009-10-01 | | Release date: | 2009-12-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Unraveling the Allosteric Mechanism of Serine Protease Inhibition by an Antibody
Structure, 17, 2009
|
|
7QKD
 
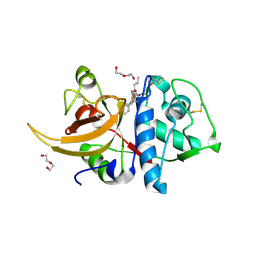 | | Crystal structure of human Cathepsin L in complex with covalently bound MG132 | | Descriptor: | ACETATE ION, Cathepsin L, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Reinke, P.Y.A, Falke, S, Lieske, J, Ewert, W, Loboda, J, Rahmani Mashhour, A, Hauser, M, Karnicar, K, Usenik, A, Lindic, N, Lach, M, Boehler, H, Beck, T, Cox, R, Chapman, H.N, Hinrichs, W, Turk, D, Guenther, S, Meents, A. | | Deposit date: | 2021-12-17 | | Release date: | 2022-12-28 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Elucidation and Antiviral Activity of Covalent Cathepsin L Inhibitors.
J.Med.Chem., 2024
|
|
2BXI
 
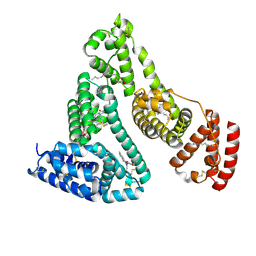 | | Human serum albumin complexed with myristate and azapropazone | | Descriptor: | AZAPROPAZONE, MYRISTIC ACID, SERUM ALBUMIN | | Authors: | Ghuman, J, Zunszain, P.A, Petitpas, I, Bhattacharya, A.A, Curry, S. | | Deposit date: | 2005-07-26 | | Release date: | 2005-09-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis of the Drug-Binding Specificity of Human Serum Albumin.
J.Mol.Biol., 353, 2005
|
|
5AIC
 
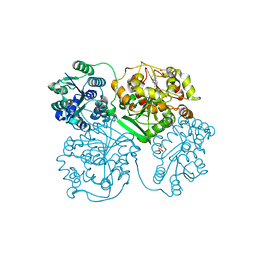 | | ligand complex structure of soluble epoxide hydrolase | | Descriptor: | 7-methyl-2H-1,4-benzothiazin-3(4H)-one, BIFUNCTIONAL EPOXIDE HYDROLASE 2, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Oster, L, Tapani, S, Xue, Y, Kack, H. | | Deposit date: | 2015-02-12 | | Release date: | 2015-05-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Successful Generation of Structural Information for Fragment-Based Drug Discovery.
Drug Discov Today, 20, 2015
|
|
2BZI
 
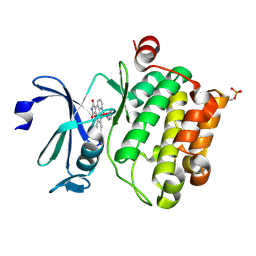 | | CRYSTAL STRUCTURE OF THE HUMAN PIM1 IN COMPLEX WITH A RUTHENIUM ORGANOMETALLIC LIGAND RU2 | | Descriptor: | PROTO-ONCOGENE SERINE THREONINE PROTEIN KINASE PIM1, RU-PYRIDOCARBAZOLE-2 | | Authors: | Debreczeni, J.E, Bullock, A, Knapp, S, von Delft, F, Sundstrom, M, Arrowsmith, C, Weigelt, J, Edwards, A. | | Deposit date: | 2005-08-18 | | Release date: | 2005-12-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the Human Pim1 in Complex with Ruthenium Organometallic Ligands
To be Published
|
|
5EUG
 
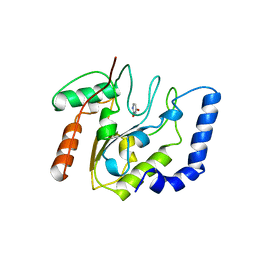 | | CRYSTALLOGRAPHIC AND ENZYMATIC STUDIES OF AN ACTIVE SITE VARIANT H187Q OF ESCHERICHIA COLI URACIL DNA GLYCOSYLASE: CRYSTAL STRUCTURES OF MUTANT H187Q AND ITS URACIL COMPLEX | | Descriptor: | PROTEIN (GLYCOSYLASE), URACIL | | Authors: | Xiao, G, Tordova, M, Drohat, A.C, Jagadeesh, J, Stivers, J.T, Gilliland, G.L. | | Deposit date: | 1998-12-27 | | Release date: | 1999-07-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of Escherichia coli uracil DNA glycosylase and its complexes with uracil and glycerol: structure and glycosylase mechanism revisited.
Proteins, 35, 1999
|
|
5ESV
 
 | | Crystal Structure of Broadly Neutralizing Antibody CH03, Isolated from Donor CH0219, in Complex with Scaffolded Trimeric HIV-1 Env V1V2 Domain from the Clade C Superinfecting Strain of Donor CAP256. | | Descriptor: | 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase,Envelope glycoprotein gp160, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gorman, J, Yang, M, Kwong, P.D. | | Deposit date: | 2015-11-17 | | Release date: | 2015-12-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.105 Å) | | Cite: | Structures of HIV-1 Env V1V2 with broadly neutralizing antibodies reveal commonalities that enable vaccine design.
Nat.Struct.Mol.Biol., 23, 2016
|
|
3E88
 
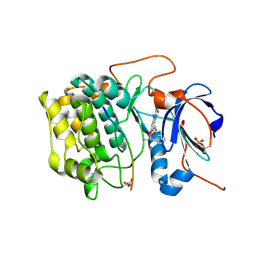 | | Crystal structures of the kinase domain of AKT2 in complex with ATP-competitive inhibitors | | Descriptor: | 4-[2-(4-amino-1,2,5-oxadiazol-3-yl)-6-{[(2R)-2-amino-3-phenylpropyl]oxy}-1-ethyl-1H-imidazo[4,5-c]pyridin-4-yl]-2-methylbut-3-yn-2-ol, Glycogen synthase kinase-3 beta peptide, RAC-beta serine/threonine-protein kinase | | Authors: | Concha, N.O, Elkins, P.A, Smallwood, A, Ward, P. | | Deposit date: | 2008-08-19 | | Release date: | 2008-10-14 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Aminofurazans as potent inhibitors of AKT kinase
Bioorg.Med.Chem.Lett., 19, 2009
|
|
2C00
 
 | |
5ES4
 
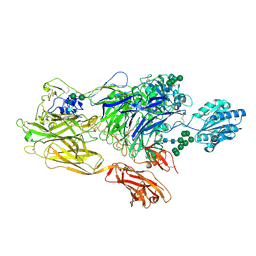 | | RE-REFINEMENT OF INTEGRIN ALPHAXBETA2 ECTODOMAIN IN THE CLOSED/BENT CONFORMATION | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Sen, M, Springer, T.A. | | Deposit date: | 2015-11-16 | | Release date: | 2016-03-02 | | Last modified: | 2022-03-23 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Leukocyte integrin alpha L beta 2 headpiece structures: The alpha I domain, the pocket for the internal ligand, and concerted movements of its loops.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5AKZ
 
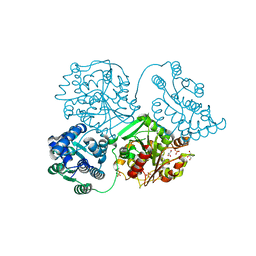 | | ligand complex structure of soluble epoxide hydrolase | | Descriptor: | 1-CYCLOHEXYL-3-METHYL-UREA, BIFUNCTIONAL EPOXIDE HYDROLASE 2, GLYCEROL, ... | | Authors: | Oster, L, Tapani, S, Xue, Y, Kack, H. | | Deposit date: | 2015-03-05 | | Release date: | 2015-05-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Successful Generation of Structural Information for Fragment-Based Drug Discovery.
Drug Discov Today, 20, 2015
|
|
5AK4
 
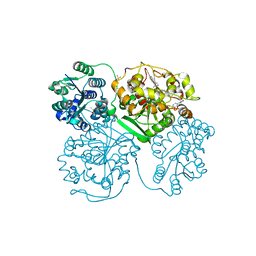 | | ligand complex structure of soluble epoxide hydrolase | | Descriptor: | 3-METHYL-4-PHENYL-1H-PYRAZOLE, BIFUNCTIONAL EPOXIDE HYDROLASE 2, DIMETHYL SULFOXIDE | | Authors: | Oster, L, Tapani, S, Xue, Y, Kack, H. | | Deposit date: | 2015-03-02 | | Release date: | 2015-05-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Successful Generation of Structural Information for Fragment-Based Drug Discovery.
Drug Discov Today, 20, 2015
|
|
4MVK
 
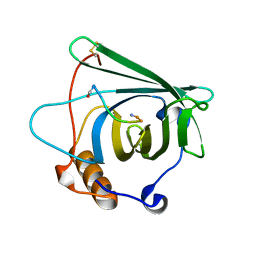 | |
