1WW2
 
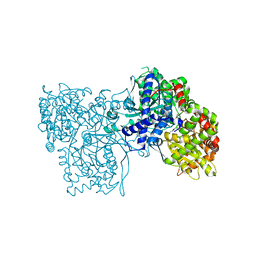 | | Crystallographic studies on two bioisosteric analogues, N-acetyl-beta-D-glucopyranosylamine and N-trifluoroacetyl-beta-D-glucopyranosylamine, potent inhibitors of muscle glycogen phosphorylase | | Descriptor: | Glycogen phosphorylase, muscle form, N-acetyl-beta-D-glucopyranosylamine, ... | | Authors: | Anagnostou, E, Kosmopoulou, M.N, Chrysina, E.D, Leonidas, D.D, Hadjiloi, T, Tiraidis, C, Zographos, S.E, Gyorgydeak, Z, Somsak, L, Docsa, T, Gergely, P, Kolisis, F.N, Oikonomakos, N.G. | | Deposit date: | 2004-12-30 | | Release date: | 2005-12-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic studies on two bioisosteric analogues, N-acetyl-beta-d-glucopyranosylamine and N-trifluoroacetyl-beta-d-glucopyranosylamine, potent inhibitors of muscle glycogen phosphorylase
Bioorg.Med.Chem., 14, 2006
|
|
1WW3
 
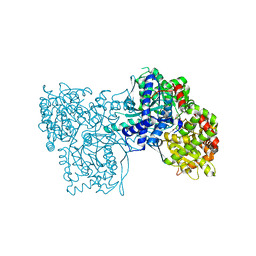 | | Crystallographic studies on two bioisosteric analogues, N-acetyl-beta-D-glucopyranosylamine and N-trifluoroacetyl-beta-D-glucopyranosylamine, potent inhibitors of muscle glycogen phosphorylase | | Descriptor: | Glycogen phosphorylase, muscle form, N-(trifluoroacetyl)-beta-D-glucopyranosylamine, ... | | Authors: | Anagnostou, E, Kosmopoulou, M.N, Chrysina, E.D, Leonidas, D.D, Hadjiloi, T, Tiraidis, C, Zographos, S.E, Gyorgydeak, Z, Somsak, L, Docsa, T, Gergely, P, Kolisis, F.N, Oikonomakos, N.G. | | Deposit date: | 2004-12-31 | | Release date: | 2005-12-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystallographic studies on two bioisosteric analogues, N-acetyl-beta-d-glucopyranosylamine and N-trifluoroacetyl-beta-d-glucopyranosylamine, potent inhibitors of muscle glycogen phosphorylase
Bioorg.Med.Chem., 14, 2006
|
|
1WW4
 
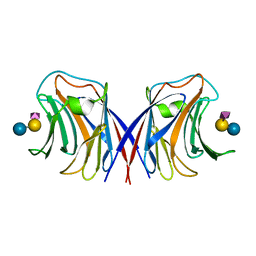 | | Agrocybe cylindracea galectin complexed with NeuAca2-3lactose | | Descriptor: | N-acetyl-alpha-neuraminic acid-(2-3)-alpha-D-galactopyranose-(1-4)-alpha-D-glucopyranose, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, galectin | | Authors: | Ban, M, Yoon, H.J, Demirkan, E, Utsumi, S, Mikami, B, Yagi, F. | | Deposit date: | 2004-12-31 | | Release date: | 2005-08-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of a Fungal Galectin from Agrocybe cylindracea for Recognizing Sialoconjugate
J.Mol.Biol., 351, 2005
|
|
1WW5
 
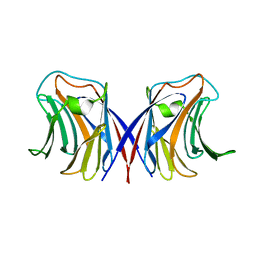 | | Agrocybe cylindracea galectin complexed with 3'-sulfonyl lactose | | Descriptor: | 3-O-sulfo-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, galectin | | Authors: | Ban, M, Yoon, H.J, Demirkan, E, Utsumi, S, Mikami, B, Yagi, F. | | Deposit date: | 2005-01-03 | | Release date: | 2005-08-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis of a Fungal Galectin from Agrocybe cylindracea for Recognizing Sialoconjugate
J.Mol.Biol., 351, 2005
|
|
1WW6
 
 | | Agrocybe cylindracea galectin complexed with lactose | | Descriptor: | beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, galectin | | Authors: | Ban, M, Yoon, H.J, Demirkan, E, Utsumi, S, Mikami, B, Yagi, F. | | Deposit date: | 2005-01-03 | | Release date: | 2005-08-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis of a Fungal Galectin from Agrocybe cylindracea for Recognizing Sialoconjugate
J.Mol.Biol., 351, 2005
|
|
1WW7
 
 | | Agrocybe cylindracea galectin (Ligand-free) | | Descriptor: | SULFATE ION, galectin | | Authors: | Ban, M, Yoon, H.J, Demirkan, E, Utsumi, S, Mikami, B, Yagi, F. | | Deposit date: | 2005-01-03 | | Release date: | 2005-08-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis of a Fungal Galectin from Agrocybe cylindracea for Recognizing Sialoconjugate
J.Mol.Biol., 351, 2005
|
|
1WW8
 
 | |
1WW9
 
 | | Crystal structure of the terminal oxygenase component of carbazole 1,9a-dioxygenase, a non-heme iron oxygenase system catalyzing the novel angular dioxygenation for carbazole and dioxin | | Descriptor: | FE (II) ION, FE2/S2 (INORGANIC) CLUSTER, terminal oxygenase component of carbazole | | Authors: | Nojiri, H, Ashikawa, Y, Noguchi, H, Nam, J.-W, Urata, M, Fujimoto, Z, Mizuno, H, Yoshida, T, Habe, H, Omori, T. | | Deposit date: | 2005-01-05 | | Release date: | 2005-08-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of the terminal oxygenase component of angular dioxygenase, carbazole 1,9a-dioxygenase
J.Mol.Biol., 351, 2005
|
|
1WWA
 
 | | NGF BINDING DOMAIN OF HUMAN TRKA RECEPTOR | | Descriptor: | PROTEIN (NERVE GROWTH FACTOR RECEPTOR TRKA) | | Authors: | Wiesmann, C, Ultsch, M.H, Bass, S.H, De Vos, A.M. | | Deposit date: | 1999-04-29 | | Release date: | 1999-07-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of the neurotrophin-binding domain of TrkA, TrkB and TrkC.
J.Mol.Biol., 290, 1999
|
|
1WWB
 
 | | LIGAND BINDING DOMAIN OF HUMAN TRKB RECEPTOR | | Descriptor: | PROTEIN (Brain Derived Neurotrophic Factor Receptor TrkB) | | Authors: | Wiesmann, C, Ultsch, M.H, Bass, S.H, De Vos, A.M. | | Deposit date: | 1999-05-03 | | Release date: | 1999-07-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of the neurotrophin-binding domain of TrkA, TrkB and TrkC.
J.Mol.Biol., 290, 1999
|
|
1WWC
 
 | | NT3 BINDING DOMAIN OF HUMAN TRKC RECEPTOR | | Descriptor: | PROTEIN (NT-3 GROWTH FACTOR RECEPTOR TRKC) | | Authors: | Ultsch, M.H, Wiesmann, C, Simmons, L.C, Henrich, J, Yang, M, Reilly, D, Bass, S.H, De Vos, A.M. | | Deposit date: | 1999-04-30 | | Release date: | 1999-07-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of the neurotrophin-binding domain of TrkA, TrkB and TrkC.
J.Mol.Biol., 290, 1999
|
|
1WWD
 
 | | NMR structure determined for MLV NC complex with RNA sequence AACAGU | | Descriptor: | 5'-R(P*AP*AP*CP*AP*GP*U)-3', Nucleoprotein p10, ZINC ION | | Authors: | Dey, A, York, D, Smalls-Mantey, A, Summers, M.F. | | Deposit date: | 2005-01-05 | | Release date: | 2005-03-22 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Composition and sequence-dependent binding of RNA to the nucleocapsid protein of Moloney murine leukemia virus(,)
Biochemistry, 44, 2005
|
|
1WWE
 
 | | NMR Structure Determined for MLV NC complex with RNA Sequence UUUUGCU | | Descriptor: | 5'-R(P*UP*UP*UP*UP*GP*CP*U)-3', Nucleoprotein p10, ZINC ION | | Authors: | Dey, A, York, D, Smalls-Mantey, A, Summers, M.F. | | Deposit date: | 2005-01-05 | | Release date: | 2005-03-22 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Composition and sequence-dependent binding of RNA to the nucleocapsid protein of Moloney murine leukemia virus(,)
Biochemistry, 44, 2005
|
|
1WWF
 
 | | NMR Structure Determined for MLV NC Complex with RNA Sequence CCUCCGU | | Descriptor: | 5'-R(P*CP*CP*UP*CP*CP*GP*U)-3', Nucleoprotein p10, ZINC ION | | Authors: | Dey, A, York, D, Smalls-Mantey, A, Summers, M.F. | | Deposit date: | 2005-01-05 | | Release date: | 2005-04-05 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Composition and sequence-dependent binding of RNA to the nucleocapsid protein of Moloney murine leukemia virus(,)
Biochemistry, 44, 2005
|
|
1WWG
 
 | | NMR Structure Determined for MLV NC Complex with RNA Sequence UAUCUG | | Descriptor: | 5'-R(P*UP*AP*UP*CP*UP*G)-3', Nucleoprotein p10, ZINC ION | | Authors: | Dey, A, York, D, Smalls-Mantey, A, Summers, M.F. | | Deposit date: | 2005-01-05 | | Release date: | 2005-04-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Composition and sequence-dependent binding of RNA to the nucleocapsid protein of Moloney murine leukemia virus(,)
Biochemistry, 44, 2005
|
|
1WWH
 
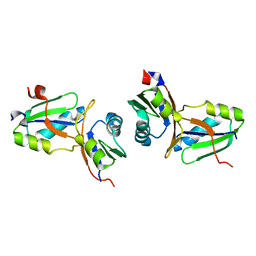 | | Crystal structure of the MPPN domain of mouse Nup35 | | Descriptor: | nucleoporin 35 | | Authors: | Handa, N, Murayama, K, Kukimoto, M, Hamana, H, Uchikubo, T, Takemoto, C, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-01-05 | | Release date: | 2005-07-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The crystal structure of mouse Nup35 reveals atypical RNP motifs and novel homodimerization of the RRM domain
J.Mol.Biol., 363, 2006
|
|
1WWI
 
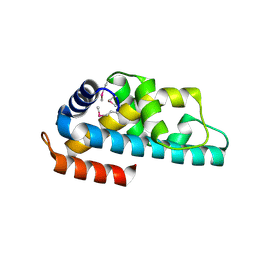 | | Crystal structure of ttk003001566 from Thermus Thermophilus HB8 | | Descriptor: | hypothetical protein TTHA1479 | | Authors: | Wang, H, Murayama, K, Terada, T, Chen, L, Liu, Z.J, Wang, B.C, Shirouzu, M, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-01-05 | | Release date: | 2005-07-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal structure of ttk003001566 from Thermus Thermophilus HB8
TO BE PUBLISHED
|
|
1WWJ
 
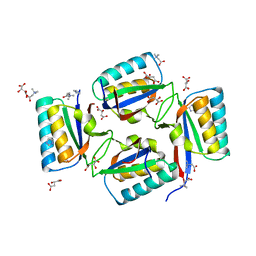 | | crystal structure of KaiB from Synechocystis sp. | | Descriptor: | Circadian clock protein kaiB, D-MALATE, IMIDAZOLE, ... | | Authors: | Hitomi, K, Oyama, T, Han, S, Arvai, A.S, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2005-01-06 | | Release date: | 2005-02-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Tetrameric architecture of the circadian clock protein KaiB. A novel interface for intermolecular interactions and its impact on the circadian rhythm.
J.Biol.Chem., 280, 2005
|
|
1WWK
 
 | |
1WWL
 
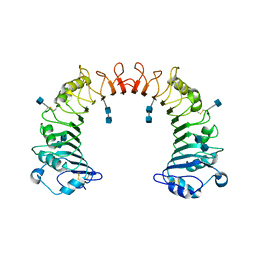 | | Crystal structure of CD14 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Monocyte differentiation antigen CD14 | | Authors: | Kim, J.-I, Lee, C.J, Jin, M.S, Lee, C.-H, Paik, S.-G, Lee, H, Lee, J.-O. | | Deposit date: | 2005-01-06 | | Release date: | 2005-02-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of CD14 and Its Implications for Lipopolysaccharide Signaling
J.Biol.Chem., 280, 2005
|
|
1WWM
 
 | | Crystal Structure of Conserved Hypothetical Protein TT2028 from an Extremely Thermophilic Bacterium Thermus thermophilus HB8 | | Descriptor: | hypothetical protein TT2028 | | Authors: | Mizohata, E, Ushikoshi-Nakayama, R, Terada, T, Murayama, K, Sakai, H, Kuramitsu, S, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-01-07 | | Release date: | 2005-07-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Crystal Structure of TT2028 from an Extremely Thermophilic Bacterium Thermus thermophilus HB8
To be Published
|
|
1WWN
 
 | | NMR Solution Structure of BmK-betaIT, an Excitatory Scorpion Toxin from Buthus martensi Karsch | | Descriptor: | Excitatory insect selective toxin 1 | | Authors: | Wu, H, Tong, X, Chen, X, Zhang, Q, Zheng, X, Zhang, N, Wu, G. | | Deposit date: | 2005-01-10 | | Release date: | 2006-01-17 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of BmK-betaIT, an excitatory scorpion beta-toxin without a 'hot spot' at the relevant position
Biochem.Biophys.Res.Commun., 349, 2006
|
|
1WWP
 
 | | Crystal structure of ttk003001694 from Thermus Thermophilus HB8 | | Descriptor: | hypothetical protein TTHA0636 | | Authors: | Wang, H, Murayama, K, Terada, T, Shirouzu, M, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-01-12 | | Release date: | 2005-07-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Crystal structure of ttk003001694 from Thermus Thermophilus HB8
TO BE PUBLISHED
|
|
1WWQ
 
 | | Solution Structure of Mouse ER | | Descriptor: | Enhancer of rudimentary homolog | | Authors: | Li, H, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-01-12 | | Release date: | 2006-01-03 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the mouse enhancer of rudimentary protein reveals a novel fold
J.Biomol.Nmr, 32, 2005
|
|
1WWR
 
 | | Crystal structure of tRNA adenosine deaminase TadA from Aquifex aeolicus | | Descriptor: | ZINC ION, tRNA adenosine deaminase TadA | | Authors: | Kuratani, M, Ishii, R, Bessho, Y, Fukunaga, R, Sengoku, T, Sekine, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-01-12 | | Release date: | 2005-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of tRNA Adenosine Deaminase (TadA) from Aquifex aeolicus
J.Biol.Chem., 280, 2005
|
|
