1I0D
 
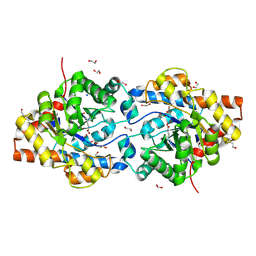 | | HIGH RESOLUTION STRUCTURE OF THE ZINC/CADMIUM-CONTAINING PHOSPHOTRIESTERASE FROM PSEUDOMONAS DIMINUTA | | Descriptor: | 1,2-ETHANEDIOL, 2-PHENYL-ETHANOL, CADMIUM ION, ... | | Authors: | Holden, H.M, Benning, M.M, Raushel, F.M, Shim, H. | | Deposit date: | 2001-01-29 | | Release date: | 2001-04-04 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | High resolution X-ray structures of different metal-substituted forms of phosphotriesterase from Pseudomonas diminuta.
Biochemistry, 40, 2001
|
|
7TZM
 
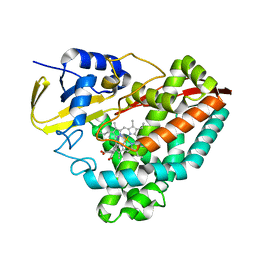 | |
7TZY
 
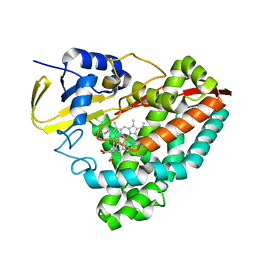 | |
4WPH
 
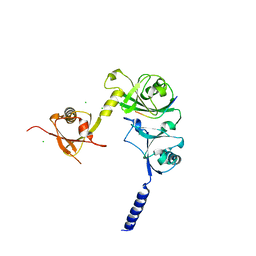 | |
1PSC
 
 | | PHOSPHOTRIESTERASE FROM PSEUDOMONAS DIMINUTA | | Descriptor: | CADMIUM ION, DIETHYL 4-METHYLBENZYLPHOSPHONATE, FORMIC ACID, ... | | Authors: | Benning, M.M, Holden, H.M. | | Deposit date: | 1995-04-25 | | Release date: | 1997-04-01 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three-dimensional structure of the binuclear metal center of phosphotriesterase.
Biochemistry, 34, 1995
|
|
1KPE
 
 | | PKCI-TRANSITION STATE ANALOG | | Descriptor: | ADENOSINE-5'-DITUNGSTATE, PROTEIN KINASE C INTERACTING PROTEIN | | Authors: | Lima, C.D, Klein, M.G, Hendrickson, W.A. | | Deposit date: | 1997-09-25 | | Release date: | 1998-03-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-based analysis of catalysis and substrate definition in the HIT protein family.
Science, 278, 1997
|
|
8CZP
 
 | |
6EZQ
 
 | | human Serum Albumin complexed with NBD-C12 fatty acid | | Descriptor: | 12-[(4-nitro-2,1,3-benzoxadiazol-7-yl)amino]dodecanoic acid, Serum albumin | | Authors: | Wenskowsky, L, Liesum, A, Schreuder, H.A. | | Deposit date: | 2017-11-16 | | Release date: | 2017-12-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Identification and Characterization of a Single High-Affinity Fatty Acid Binding Site in Human Serum Albumin.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
7TZN
 
 | |
3WT6
 
 | | A mixed population of antagonist and agonist binding conformers in a single crystal explains partial agonism against vitamin D receptor: Active vitamin D analogues with 22R-alkyl group | | Descriptor: | (1R,3R,7E,17beta)-17-[(2R,3R)-3-butyl-6-hydroxy-6-methylheptan-2-yl]-2-methylidene-9,10-secoestra-5,7-diene-1,3-diol, Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 receptor | | Authors: | Anami, Y, Itoh, T, Yamamoto, K. | | Deposit date: | 2014-04-08 | | Release date: | 2014-06-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Mixed Population of Antagonist and Agonist Binding Conformers in a Single Crystal Explains Partial Agonism against Vitamin D Receptor: Active Vitamin D Analogues with 22R-Alkyl Group.
J.Med.Chem., 57, 2014
|
|
1KS8
 
 | | The structure of Endoglucanase from termite, Nasutitermes takasagoensis, at pH 2.5. | | Descriptor: | Endo-b-1,4-glucanase, SULFATE ION | | Authors: | Khademi, S, Guarino, L.A, Watanabe, H, Tokuda, G, Meyer, E.F. | | Deposit date: | 2002-01-11 | | Release date: | 2003-01-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of an endoglucanase from termite, Nasutitermes takasagoensis.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
4X5A
 
 | |
1KW1
 
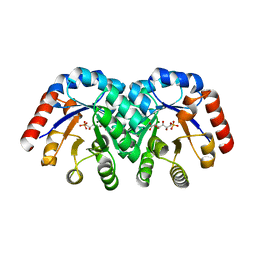 | | Crystal Structure of 3-Keto-L-Gulonate 6-Phosphate Decarboxylase with bound L-gulonate 6-phosphate | | Descriptor: | 3-Keto-L-Gulonate 6-Phosphate Decarboxylase, L-GULURONIC ACID 6-PHOSPHATE, MAGNESIUM ION | | Authors: | Wise, E, Yew, W.S, Babbitt, P.C, Gerlt, J.A, Rayment, I. | | Deposit date: | 2002-01-28 | | Release date: | 2002-04-15 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Homologous (beta/alpha)8-barrel enzymes that catalyze unrelated reactions: orotidine 5'-monophosphate decarboxylase and 3-keto-L-gulonate 6-phosphate decarboxylase.
Biochemistry, 41, 2002
|
|
2HYQ
 
 | |
1KU7
 
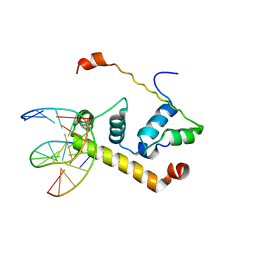 | | Crystal Structure of Thermus aquatics RNA Polymerase SigmaA Subunit Region 4 Bound to-35 Element DNA | | Descriptor: | 5'-D(*CP*CP*TP*TP*GP*AP*CP*AP*AP*AP*G)-3', 5'-D(*CP*CP*TP*TP*TP*GP*TP*CP*AP*AP*G)-3', sigma factor sigA | | Authors: | Campbell, E.A, Muzzin, O, Chlenov, M, Sun, J.L, Olson, C.A, Weinman, O, Trester-Zedlitz, M.L, Darst, S.A. | | Deposit date: | 2002-01-21 | | Release date: | 2002-03-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the bacterial RNA polymerase promoter specificity sigma subunit.
Mol.Cell, 9, 2002
|
|
1IMC
 
 | |
1KU3
 
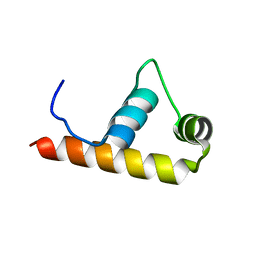 | | Crystal Structure of Thermus aquaticus RNA Polymerase Sigma Subunit Fragment, Region 4 | | Descriptor: | sigma factor sigA | | Authors: | Campbell, E.A, Muzzin, O, Chlenov, M, Sun, J.L, Olson, C.A, Weinman, O, Trester-Zedlitz, M.L, Darst, S.A. | | Deposit date: | 2002-01-21 | | Release date: | 2002-04-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the bacterial RNA polymerase promoter specificity sigma subunit.
Mol.Cell, 9, 2002
|
|
8E1A
 
 | | Structure-based study to overcome cross-reactivity of novel androgen receptor inhibitors | | Descriptor: | 1,2-ETHANEDIOL, 4-[4-(3-fluoro-2-methoxyphenyl)-1,3-thiazol-2-yl]morpholine, Androgen receptor | | Authors: | Lallous, N, Li, H, Radaeva, M, Dalal, K, Leblanc, E, Ban, F, Ciesielski, F, Chow, B, Morin, M, Singh, K, Rennie, P.S, Cherkasov, A. | | Deposit date: | 2022-08-10 | | Release date: | 2022-09-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure-Based Study to Overcome Cross-Reactivity of Novel Androgen Receptor Inhibitors.
Cells, 11, 2022
|
|
2HZ4
 
 | | Abl kinase domain unligated and in complex with tetrahydrostaurosporine | | Descriptor: | 1,2,3,4-TETRAHYDROGEN-STAUROSPORINE, Proto-oncogene tyrosine-protein kinase ABL1 | | Authors: | Cowan-Jacob, S.W, Fendrich, G, Liebetanz, J, Fabbro, D, Manley, P. | | Deposit date: | 2006-08-08 | | Release date: | 2007-01-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural biology contributions to the discovery of drugs to treat chronic myelogenous leukaemia.
ACTA CRYSTALLOGR.,SECT.D, 63, 2007
|
|
2HYR
 
 | | Crystal structure of a complex of griffithsin with maltose | | Descriptor: | 1,2-ETHANEDIOL, Griffithsin, MAGNESIUM ION, ... | | Authors: | Ziolkowska, N.E, Wlodawer, A. | | Deposit date: | 2006-08-07 | | Release date: | 2007-04-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Crystallographic, thermodynamic, and molecular modeling studies of the mode of binding of oligosaccharides to the potent antiviral protein griffithsin.
Proteins, 67, 2007
|
|
7V58
 
 | | Structural insights into the substrate selectivity of acyl-CoA transferase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-amino-3-ketobutyrate coenzyme A ligase, GLYCINE, ... | | Authors: | Chang, H.Y, Ko, T.P. | | Deposit date: | 2021-08-16 | | Release date: | 2021-12-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural insights into the substrate selectivity of alpha-oxoamine synthases from marine Vibrio sp. QWI-06.
Colloids Surf B Biointerfaces, 210, 2022
|
|
7V5I
 
 | |
1KPF
 
 | | PKCI-SUBSTRATE ANALOG | | Descriptor: | ADENOSINE MONOPHOSPHATE, PROTEIN KINASE C INTERACTING PROTEIN | | Authors: | Lima, C.D, Klein, M.G, Hendrickson, W.A. | | Deposit date: | 1997-09-25 | | Release date: | 1998-03-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-based analysis of catalysis and substrate definition in the HIT protein family.
Science, 278, 1997
|
|
5KF1
 
 | | X-ray structure of a glucosamine N-Acetyltransferase from Clostridium acetobutylicum, apo form, pH 5 | | Descriptor: | 1,2-ETHANEDIOL, ACETYL COENZYME *A, COENZYME A, ... | | Authors: | Holden, H.M, Thoden, J.B, Dopkins, B.J, tipton, P.A. | | Deposit date: | 2016-06-11 | | Release date: | 2016-07-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Studies on a Glucosamine/Glucosaminide N-Acetyltransferase.
Biochemistry, 55, 2016
|
|
1KUH
 
 | | ZINC PROTEASE FROM STREPTOMYCES CAESPITOSUS | | Descriptor: | CALCIUM ION, ZINC ION, ZINC PROTEASE | | Authors: | Kurisu, G, Kinoshita, T, Sugimoto, A, Nagara, A, Kai, Y, Kasai, N, Harada, S. | | Deposit date: | 1996-02-22 | | Release date: | 1997-03-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of the zinc endoprotease from Streptomyces caespitosus.
J.Biochem.(Tokyo), 121, 1997
|
|
