1WPT
 
 | | Crystal Structure of HutP, an RNA binding anti-termination protein | | Descriptor: | Hut operon positive regulatory protein | | Authors: | Kumarevel, T, Mizuno, H, Kumar, P.K.R. | | Deposit date: | 2004-09-13 | | Release date: | 2005-08-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Characterization of the metal ion binding site in the anti-terminator protein, HutP, of Bacillus subtilis
Nucleic Acids Res., 33, 2005
|
|
1WPU
 
 | |
1WPV
 
 | | Crystal Structure of Activated Binary complex of HutP, an RNA binding anti-termination protein | | Descriptor: | HISTIDINE, Hut operon positive regulatory protein, MAGNESIUM ION | | Authors: | Kumarevel, T.S, Mizuno, H, Kumar, P.K.R. | | Deposit date: | 2004-09-14 | | Release date: | 2005-03-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis of HutP-mediated anti-termination and roles of the Mg2+ ion and L-histidine ligand.
Nature, 434, 2005
|
|
1WPW
 
 | | Crystal Structure of IPMDH from Sulfolobus tokodaii | | Descriptor: | 3-isopropylmalate dehydrogenase, MAGNESIUM ION | | Authors: | Hirose, R, Sakurai, M, Suzuki, T, Moriyama, H, Sato, T, Yamagishi, A, Oshima, T, Tanaka, N. | | Deposit date: | 2004-09-14 | | Release date: | 2004-10-05 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of IPMDH from Sulfolobus tokodaii
To be Published
|
|
1WPX
 
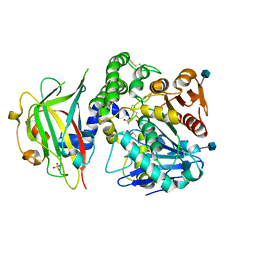 | | Crystal structure of carboxypeptidase Y inhibitor complexed with the cognate proteinase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Carboxypeptidase Y, Carboxypeptidase Y inhibitor, ... | | Authors: | Mima, J, Hayashida, M, Fujii, T, Narita, Y, Hayashi, R, Ueda, M, Hata, Y. | | Deposit date: | 2004-09-14 | | Release date: | 2005-03-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of the carboxypeptidase y inhibitor i(c) in complex with the cognate proteinase reveals a novel mode of the proteinase-protein inhibitor interaction
J.Mol.Biol., 346, 2005
|
|
1WPY
 
 | |
1WQ1
 
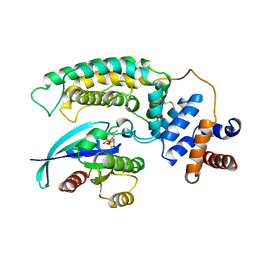 | | RAS-RASGAP COMPLEX | | Descriptor: | ALUMINUM FLUORIDE, GUANOSINE-5'-DIPHOSPHATE, H-RAS, ... | | Authors: | Scheffzek, K, Ahmadian, M.R, Kabsch, W, Wiesmueller, L, Lautwein, A, Schmitz, F, Wittinghofer, A. | | Deposit date: | 1997-07-03 | | Release date: | 1998-07-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Ras-RasGAP complex: structural basis for GTPase activation and its loss in oncogenic Ras mutants.
Science, 277, 1997
|
|
1WQ2
 
 | | Neutron Crystal Structure Of Dissimilatory Sulfite Reductase D (DsrD) | | Descriptor: | Protein dsvD, SULFATE ION | | Authors: | Chatake, T, Mizuno, N, Voordouw, G, Higuchi, Y, Arai, S, Tanaka, I, Niimura, N. | | Deposit date: | 2004-09-19 | | Release date: | 2005-09-19 | | Last modified: | 2023-10-25 | | Method: | NEUTRON DIFFRACTION (2.4 Å) | | Cite: | Crystallization and preliminary neutron analysis of the dissimilatory sulfite reductase D (DsrD) protein from the sulfate-reducing bacterium Desulfovibrio vulgaris.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1WQ3
 
 | | Escherichia coli tyrosyl-tRNA synthetase mutant complexed with 3-iodo-L-tyrosine | | Descriptor: | 3-IODO-TYROSINE, Tyrosyl-tRNA synthetase | | Authors: | Kobayashi, T, Sakamoto, K, Nureki, O, Takimura, T, Kamata, K, Sekine, R, Nishimura, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-09-20 | | Release date: | 2005-01-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of nonnatural amino acid recognition by an engineered aminoacyl-tRNA synthetase for genetic code expansion
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
1WQ4
 
 | | Escherichia coli tyrosyl-tRNA synthetase mutant complexed with L-tyrosine | | Descriptor: | TYROSINE, Tyrosyl-tRNA synthetase | | Authors: | Kobayashi, T, Sakamoto, K, Nureki, O, Takimura, T, Kamata, K, Sekine, R, Nishimura, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-09-20 | | Release date: | 2005-01-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of nonnatural amino acid recognition by an engineered aminoacyl-tRNA synthetase for genetic code expansion
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
1WQ5
 
 | | Crystal structure of tryptophan synthase alpha-subunit from Escherichia coli | | Descriptor: | GLYCEROL, SULFATE ION, Tryptophan synthase alpha chain | | Authors: | Nishio, K, Morimoto, Y, Ishizuka, M, Ogasahara, K, Yutani, K, Tsukihara, T, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-09-22 | | Release date: | 2005-02-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Conformational Changes in the alpha-Subunit Coupled to Binding of the beta(2)-Subunit of Tryptophan Synthase from Escherichia coli: Crystal Structure of the Tryptophan Synthase alpha-Subunit Alon
Biochemistry, 44, 2005
|
|
1WQ6
 
 | | The tetramer structure of the nervy homolgy two (NHR2) domain of AML1-ETO is critical for AML1-ETO'S activity | | Descriptor: | AML1-ETO | | Authors: | Liu, Y, Cheney, M.D, Chruszcz, M, Lukasik, S.M, Hartman, K.L, Laue, T.M, Dauter, Z, Minor, W, Speck, N.A, Bushweller, J.H. | | Deposit date: | 2004-09-23 | | Release date: | 2005-10-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The tetramer structure of the Nervy homology two domain, NHR2, is critical for AML1/ETO's activity
Cancer Cell, 9, 2006
|
|
1WQ7
 
 | |
1WQ8
 
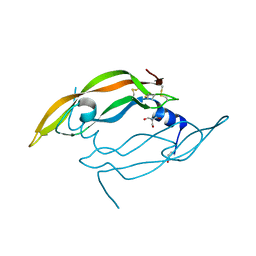 | | Crystal structure of Vammin, a VEGF-F from a snake venom | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Vascular endothelial growth factor toxin | | Authors: | Suto, K, Yamazaki, Y, Morita, T, Mizuno, H. | | Deposit date: | 2004-09-23 | | Release date: | 2004-12-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of novel vascular endothelial growth factors (VEGF) from snake venoms: insight into selective VEGF binding to kinase insert domain-containing receptor but not to fms-like tyrosine kinase-1.
J.Biol.Chem., 280, 2005
|
|
1WQ9
 
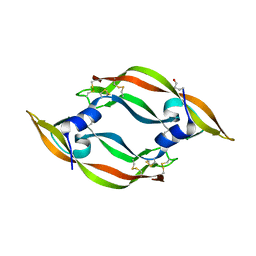 | | Crystal structure of VR-1, a VEGF-F from a snake venom | | Descriptor: | Vascular endothelial growth factor | | Authors: | Suto, K, Yamazaki, Y, Morita, T, Mizuno, H. | | Deposit date: | 2004-09-24 | | Release date: | 2004-12-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of novel vascular endothelial growth factors (VEGF) from snake venoms: insight into selective VEGF binding to kinase insert domain-containing receptor but not to fms-like tyrosine kinase-1.
J.Biol.Chem., 280, 2005
|
|
1WQA
 
 | |
1WQB
 
 | |
1WQC
 
 | | An unusual fold for potassium channel blockers : NMR structure of three toxins from the scorpion Opisthacanthus madagascariensis | | Descriptor: | OmTx1 | | Authors: | Chagot, B, Pimentel, C, Dai, L, Pil, J, Tytgat, J, Nakajima, T, Corzo, G, Darbon, H, Ferrat, G. | | Deposit date: | 2004-09-27 | | Release date: | 2005-01-18 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | An unusual fold for potassium channel blockers: NMR structure of three toxins from the scorpion Opisthacanthus madagascariensis
Biochem.J., 388, 2005
|
|
1WQD
 
 | | An unusual fold for potassium channel blockers: NMR structure of three toxins from the scorpion Opisthacanthus madagascariensis | | Descriptor: | OmTx2 | | Authors: | Chagot, B, Pimentel, C, Dai, L, Pil, J, Tytgat, J, Nakajima, T, Corzo, G, Darbon, H, Ferrat, G. | | Deposit date: | 2004-09-28 | | Release date: | 2005-01-18 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | An unusual fold for potassium channel blockers: NMR structure of three toxins from the scorpion Opisthacanthus madagascariensis
Biochem.J., 388, 2005
|
|
1WQE
 
 | | An unusual fold for potassium channel blockers: NMR structure of three toxins from the scorpion Opisthacanthus madagascariensis | | Descriptor: | OmTx3 | | Authors: | Chagot, B, Pimentel, C, Dai, L, Pil, J, Tytgat, J, Nakajima, T, Corzo, G, Darbon, H, Ferrat, G. | | Deposit date: | 2004-09-28 | | Release date: | 2005-01-18 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | An unusual fold for potassium channel blockers: NMR structure of three toxins from the scorpion Opisthacanthus madagascariensis
Biochem.J., 388, 2005
|
|
1WQF
 
 | | Crystal structure of Ribosome recycling factor from Mycobacterium Tuberculosis | | Descriptor: | CADMIUM ION, Ribosome recycling factor | | Authors: | Saikrishnan, K, Kalapala, S.K, Varshney, U, Vijayan, M. | | Deposit date: | 2004-09-28 | | Release date: | 2005-02-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | X-ray structural studies of Mycobacterium tuberculosis RRF and a comparative study of RRFs of known structure. Molecular plasticity and biological implications
J.Mol.Biol., 345, 2005
|
|
1WQG
 
 | | Crystal structure of ribosome recycling factor from Mycobacterium Tuberculosis | | Descriptor: | CADMIUM ION, Ribosome recycling factor | | Authors: | Saikrishnan, K, Kalapala, S.K, Varshney, U, Vijayan, M. | | Deposit date: | 2004-09-29 | | Release date: | 2005-02-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | X-ray structural studies of Mycobacterium tuberculosis RRF and a comparative study of RRFs of known structure. Molecular plasticity and biological implications
J.Mol.Biol., 345, 2005
|
|
1WQH
 
 | | Crystal structure of ribosome recycling factor from Mycobacterium tuberculosis | | Descriptor: | CADMIUM ION, Ribosome recycling factor | | Authors: | Saikrishnan, K, Kalapala, S.K, Varshney, U, Vijayan, M. | | Deposit date: | 2004-09-29 | | Release date: | 2005-02-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | X-ray structural studies of Mycobacterium Tuberculosis RRF and a comparative study of RRFS of known structure. Molecular plasticity and biological implications
J.Mol.Biol., 345, 2005
|
|
1WQJ
 
 | | Structural Basis for the Regulation of Insulin-Like Growth Factors (IGFs) by IGF Binding Proteins (IGFBPs) | | Descriptor: | Insulin-like growth factor IB, Insulin-like growth factor binding protein 4 | | Authors: | Siwanowicz, I, Popowicz, G.M, Wisniewska, M, Huber, R, Kuenkele, K.P, Lang, K, Engh, R.A, Holak, T.A. | | Deposit date: | 2004-09-29 | | Release date: | 2005-03-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for the regulation of insulin-like growth factors by IGF binding proteins
Structure, 13, 2005
|
|
1WQK
 
 | | Solution structure of APETx1, a specific peptide inhibitor of human Ether-a-go-go-related gene potassium channels from the venom of the sea anemone Anthopleura elegantissima: a new fold for an HERG toxin | | Descriptor: | Toxin APETx1 | | Authors: | Chagot, B, Diochot, S, Pimentel, C, Lazdunski, M, Darbon, H. | | Deposit date: | 2004-09-30 | | Release date: | 2005-03-08 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Solution structure of APETx1 from the sea anemone Anthopleura elegantissima: A new fold for an HERG toxin
Proteins, 59, 2005
|
|
