1AO0
 
 | |
4HME
 
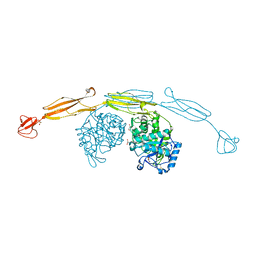 | | Crystal structure of cold-adapted chitinase from Moritella marina with a reaction product - NAG2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase 60, GLYCEROL, ... | | Authors: | Malecki, P.H, Vorgias, C.E, Raczynska, J.E, Rypniewski, W. | | Deposit date: | 2012-10-18 | | Release date: | 2013-05-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structure of a complete four-domain chitinase from Moritella marina, a marine psychrophilic bacterium
Acta Crystallogr.,Sect.D, 69, 2013
|
|
1AEL
 
 | |
1AMD
 
 | |
1A7B
 
 | |
1A2O
 
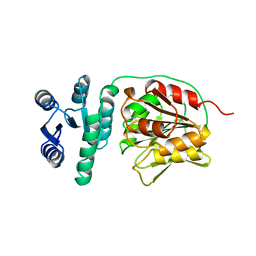 | | STRUCTURAL BASIS FOR METHYLESTERASE CHEB REGULATION BY A PHOSPHORYLATION-ACTIVATED DOMAIN | | Descriptor: | CHEB METHYLESTERASE | | Authors: | Djordjevic, S, Goudreau, P.N, Xu, Q, Stock, A.M, West, A.H. | | Deposit date: | 1998-01-06 | | Release date: | 1998-04-29 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for methylesterase CheB regulation by a phosphorylation-activated domain.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1A2D
 
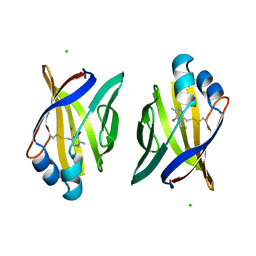 | | PYRIDOXAMINE MODIFIED MURINE ADIPOCYTE LIPID BINDING PROTEIN | | Descriptor: | ADIPOCYTE LIPID BINDING PROTEIN, CHLORIDE ION | | Authors: | Ory, J, Mazhary, A, Kuang, H, Davies, R, Distefano, M, Banaszak, L. | | Deposit date: | 1997-12-29 | | Release date: | 1998-07-01 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural characterization of two synthetic catalysts based on adipocyte lipid-binding protein.
Protein Eng., 11, 1998
|
|
1A7U
 
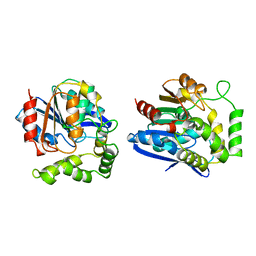 | | CHLOROPEROXIDASE T | | Descriptor: | CHLOROPEROXIDASE T | | Authors: | Hofmann, B, Toelzer, S, Pelletier, I, Altenbuchner, J, Van Pee, K.-H, Hecht, H.-J. | | Deposit date: | 1998-03-17 | | Release date: | 1998-06-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural investigation of the cofactor-free chloroperoxidases.
J.Mol.Biol., 279, 1998
|
|
6YAO
 
 | | Crystal structure of ZmCKO4a in complex with inhibitor 1-[2-(2-Hydroxy-ethyl)-phenyl]-3-(3-trifluoromethoxy-phenyl)-urea | | Descriptor: | 1,2-ETHANEDIOL, 1-[2-(2-Hydroxy-ethyl)-phenyl]-3-(3-trifluoromethoxy-phenyl)-urea, Cytokinin dehydrogenase 4, ... | | Authors: | Kopecny, D, Briozzo, P, Morera, S. | | Deposit date: | 2020-03-12 | | Release date: | 2020-10-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Diphenylurea-derived cytokinin oxidase/dehydrogenase inhibitors for biotechnology and agriculture.
J.Exp.Bot., 72, 2021
|
|
1AZF
 
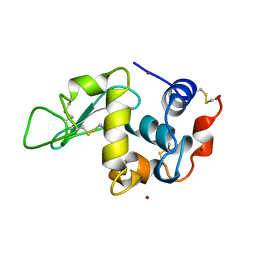 | | CHICKEN EGG WHITE LYSOZYME CRYSTAL GROWN IN BROMIDE SOLUTION | | Descriptor: | BROMIDE ION, LYSOZYME | | Authors: | Lim, K, Nadarajah, A, Forsythe, E.L, Pusey, M.L. | | Deposit date: | 1997-11-17 | | Release date: | 1998-02-25 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Locations of bromide ions in tetragonal lysozyme crystals.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
5SCK
 
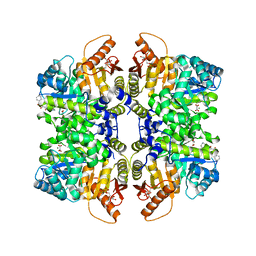 | | Structure of liver pyruvate kinase in complex with anthraquinone derivative 42 | | Descriptor: | 1,2-dihydroxy-3-(piperazine-1-sulfonyl)anthracene-9,10-dione, 1,6-di-O-phosphono-beta-D-fructofuranose, MAGNESIUM ION, ... | | Authors: | Lulla, A, Foller, A, Nain-Perez, A, Grotli, M, Brear, P, Hyvonen, M. | | Deposit date: | 2021-12-01 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.717 Å) | | Cite: | Anthraquinone derivatives as ADP-competitive inhibitors of liver pyruvate kinase.
Eur.J.Med.Chem., 234, 2022
|
|
1B0L
 
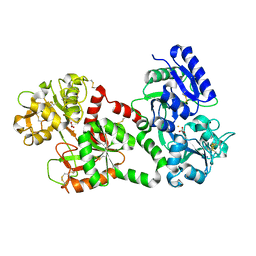 | | RECOMBINANT HUMAN DIFERRIC LACTOFERRIN | | Descriptor: | CARBONATE ION, FE (III) ION, PROTEIN (LACTOFERRIN) | | Authors: | Baker, E.N, Jameson, G.B, Sun, X. | | Deposit date: | 1998-11-11 | | Release date: | 1999-11-18 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of recombinant human lactoferrin expressed in Aspergillus awamori.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
5DXI
 
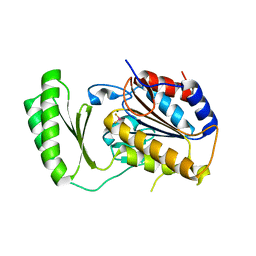 | | Structure of C. albicans Trehalose-6-phosphate phosphatase C-terminal domain | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, alpha-D-glucopyranose-(1-1)-alpha-D-glucopyranose, ... | | Authors: | Miao, Y, Brennan, R.G. | | Deposit date: | 2015-09-23 | | Release date: | 2016-06-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of trehalose-6-phosphate phosphatase from pathogenic fungi reveal the mechanisms of substrate recognition and catalysis.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
1B2M
 
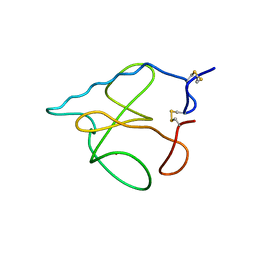 | | THREE-DIMENSIONAL STRUCTURE OF RIBONULCEASE T1 COMPLEXED WITH AN ISOSTERIC PHOSPHONATE ANALOGUE OF GPU: ALTERNATE SUBSTRATE BINDING MODES AND CATALYSIS. | | Descriptor: | 5'-R(*GP*(U34))-3', RIBONUCLEASE T1 | | Authors: | Arni, R.K, Watanabe, L, Ward, R.J, Kreitman, R.J, Kumar, K, Walz Jr, F.G. | | Deposit date: | 1998-11-27 | | Release date: | 1999-03-25 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three-dimensional structure of ribonuclease T1 complexed with an isosteric phosphonate substrate analogue of GpU: alternate substrate binding modes and catalysis.
Biochemistry, 38, 1999
|
|
1B2W
 
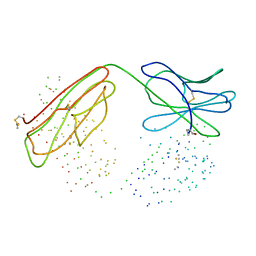 | | COMPARISON OF THE THREE-DIMENSIONAL STRUCTURES OF A HUMANIZED AND A CHIMERIC FAB OF AN ANTI-GAMMA-INTERFERON ANTIBODY | | Descriptor: | PROTEIN (ANTIBODY (HEAVY CHAIN)), PROTEIN (ANTIBODY (LIGHT CHAIN)) | | Authors: | Fan, Z, Shan, L, Goldsteen, B.Z, Guddat, L.W, Thakur, A, Landolfi, N.F, Co, M.S, Vasquez, M, Queen, C, Ramsland, P.A, Edmundson, A.B. | | Deposit date: | 1998-12-01 | | Release date: | 1999-05-06 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Comparison of the three-dimensional structures of a humanized and a chimeric Fab of an anti-gamma-interferon antibody.
J.Mol.Recog., 12, 1999
|
|
1B33
 
 | | STRUCTURE OF LIGHT HARVESTING COMPLEX OF ALLOPHYCOCYANIN ALPHA AND BETA CHAINS/CORE-LINKER COMPLEX AP*LC7.8 | | Descriptor: | ALLOPHYCOCYANIN, ALPHA CHAIN, BETA CHAIN, ... | | Authors: | Reuter, W, Wiegand, G, Huber, R, Than, M.E. | | Deposit date: | 1998-12-15 | | Release date: | 1999-02-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural analysis at 2.2 A of orthorhombic crystals presents the asymmetry of the allophycocyanin-linker complex, AP.LC7.8, from phycobilisomes of Mastigocladus laminosus.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1B4B
 
 | | STRUCTURE OF THE OLIGOMERIZATION DOMAIN OF THE ARGININE REPRESSOR FROM BACILLUS STEAROTHERMOPHILUS | | Descriptor: | ARGININE, ARGININE REPRESSOR | | Authors: | Ni, J, Sakanyan, V, Charlier, D, Glansdorff, N, Van Duyne, G.D. | | Deposit date: | 1998-12-18 | | Release date: | 1999-06-15 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the arginine repressor from Bacillus stearothermophilus.
Nat.Struct.Biol., 6, 1999
|
|
5SCI
 
 | | Structure of liver pyruvate kinase in complex with anthraquinone derivative 105 | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, MAGNESIUM ION, OXALATE ION, ... | | Authors: | Lulla, A, Foller, A, Nain-Perez, A, Grotli, M, Brear, P, Hyvonen, M. | | Deposit date: | 2021-12-01 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.155 Å) | | Cite: | Anthraquinone derivatives as ADP-competitive inhibitors of liver pyruvate kinase.
Eur.J.Med.Chem., 234, 2022
|
|
1B4J
 
 | | COMPARISON OF THE THREE-DIMENSIONAL STRUCTURES OF A HUMANIZED AND A CHIMERIC FAB OF AN ANTI-GAMMA-INTERFERON ANTIBODY | | Descriptor: | ANTIBODY | | Authors: | Fan, Z, Shan, L, Goldsteen, B.Z, Guddat, L.W, Thakur, A, Landolfi, N.F, Co, M.S, Vasques, M, Queen, C, Ramsland, P.A, Edmundson, A.B. | | Deposit date: | 1998-12-22 | | Release date: | 1999-06-15 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Comparison of the three-dimensional structures of a humanized and a chimeric Fab of an anti-gamma-interferon antibody.
J.Mol.Recog., 12, 1999
|
|
5SCF
 
 | | Structure of liver pyruvate kinase in complex with anthraquinone derivative 99 | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, MAGNESIUM ION, N-(3-hydroxy-9,10-dioxo-9,10-dihydroanthracene-2-sulfonyl)glycine, ... | | Authors: | Lulla, A, Foller, A, Nain-Perez, A, Grotli, M, Brear, P, Hyvonen, M. | | Deposit date: | 2021-12-01 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.185 Å) | | Cite: | Anthraquinone derivatives as ADP-competitive inhibitors of liver pyruvate kinase.
Eur.J.Med.Chem., 234, 2022
|
|
5SDT
 
 | | Structure of liver pyruvate kinase in complex with anthraquinone derivative 15 | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, MAGNESIUM ION, N-(3,4-dihydroxy-9,10-dioxo-9,10-dihydroanthracene-2-sulfonyl)-beta-alanine, ... | | Authors: | Lulla, A, Foller, A, Nain-Perez, A, Grotli, M, Brear, P, Hyvonen, M. | | Deposit date: | 2022-01-20 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.944 Å) | | Cite: | Anthraquinone derivatives as ADP-competitive inhibitors of liver pyruvate kinase.
Eur.J.Med.Chem., 234, 2022
|
|
1ATO
 
 | | THE STRUCTURE OF THE ISOLATED, CENTRAL HAIRPIN OF THE HDV ANTIGENOMIC RIBOZYME, NMR, 10 STRUCTURES | | Descriptor: | RNA (5'-R(*GP*GP*CP*AP*CP*CP*UP*CP*CP*UP*CP*GP*CP*GP*GP*UP*GP*CP*C)-3') | | Authors: | Kolk, M.H, Heus, H.A, Hilbers, C.W. | | Deposit date: | 1997-08-14 | | Release date: | 1997-11-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The structure of the isolated, central hairpin of the HDV antigenomic ribozyme: novel structural features and similarity of the loop in the ribozyme and free in solution.
EMBO J., 16, 1997
|
|
5SCG
 
 | | Structure of liver pyruvate kinase in complex with anthraquinone derivative 101 | | Descriptor: | (3R)-1-(3-hydroxy-9,10-dioxo-9,10-dihydroanthracene-2-sulfonyl)piperidine-3-carboxylic acid, 1,6-di-O-phosphono-beta-D-fructofuranose, MAGNESIUM ION, ... | | Authors: | Lulla, A, Foller, A, Nain-Perez, A, Grotli, M, Brear, P, Hyvonen, M. | | Deposit date: | 2021-12-01 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.937 Å) | | Cite: | Anthraquinone derivatives as ADP-competitive inhibitors of liver pyruvate kinase.
Eur.J.Med.Chem., 234, 2022
|
|
5SCJ
 
 | | Structure of liver pyruvate kinase in complex with anthraquinone derivative 106 | | Descriptor: | (2R)-2-hydroxy-2-{2-[4-(3-hydroxy-9,10-dioxo-9,10-dihydroanthracene-2-sulfonyl)piperazin-1-yl]-2-oxoethyl}butanedioic acid, 1,6-di-O-phosphono-beta-D-fructofuranose, MAGNESIUM ION, ... | | Authors: | Lulla, A, Foller, A, Nain-Perez, A, Grotli, M, Brear, P, Hyvonen, M. | | Deposit date: | 2021-12-01 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.354 Å) | | Cite: | Anthraquinone derivatives as ADP-competitive inhibitors of liver pyruvate kinase.
Eur.J.Med.Chem., 234, 2022
|
|
1B7A
 
 | | STRUCTURE OF THE PHOSPHATIDYLETHANOLAMINE-BINDING PROTEIN FROM BOVINE BRAIN | | Descriptor: | PHOSPHATIDYLETHANOLAMINE-BINDING PROTEIN, PHOSPHORIC ACID MONO-(2-AMINO-ETHYL) ESTER | | Authors: | Serre, L, Vallee, B, Bureaud, N, Schoentgen, F, Zelwer, C. | | Deposit date: | 1999-01-21 | | Release date: | 1999-01-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of the phosphatidylethanolamine-binding protein from bovine brain: a novel structural class of phospholipid-binding proteins.
Structure, 6, 1998
|
|
