7OOL
 
 | | Crystal structure of a Candidatus photodesmus katoptron thioredoxin chimera containing an ancestral loop | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Gavira, J.A, Ibarra-Molero, B, Gamiz-Arco, G, Risso, V, Sanchez-Ruiz, J.M. | | Deposit date: | 2021-05-28 | | Release date: | 2021-11-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Combining Ancestral Reconstruction with Folding-Landscape Simulations to Engineer Heterologous Protein Expression.
J.Mol.Biol., 433, 2021
|
|
2Q5Z
 
 | | Crystal structure of iMazG from Vibrio DAT 722: Ntag-iMazG (P43212) | | Descriptor: | GLYCEROL, Hypothetical protein, MAGNESIUM ION | | Authors: | Robinson, A, Guilfoyle, A.P, Harrop, S.J, Boucher, Y, Stokes, H.W, Curmi, P.M.G, Mabbutt, B.C. | | Deposit date: | 2007-06-04 | | Release date: | 2007-10-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A putative house-cleaning enzyme encoded within an integron array: 1.8 A crystal structure defines a new MazG subtype.
Mol.Microbiol., 66, 2007
|
|
7ZK6
 
 | | ABCB1 L335C mutant (mABCB1) in the outward facing state bound to 2 molecules of AAC | | Descriptor: | (4S,11S,18S)-4-[[(2,4-dinitrophenyl)disulfanyl]methyl]-11,18-dimethyl-6,13,20-trithia-3,10,17,22,23,24-hexazatetracyclo[17.2.1.1^{5,8}.1^{12,15}]tetracosa-1(21),5(24),7,12(23),14,19(22)-hexaene-2,9,16-trione, (4~{S},11~{S},18~{S})-4,11-dimethyl-18-(sulfanylmethyl)-6,13,20-trithia-3,10,17,22,23,24-hexazatetracyclo[17.2.1.1^{5,8}.1^{12,15}]tetracosa-1(21),5(24),7,12(23),14,19(22)-hexaene-2,9,16-trione, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Parey, K, Januliene, D, Gewering, T, Moeller, A. | | Deposit date: | 2022-04-12 | | Release date: | 2023-04-26 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Tracing the substrate translocation mechanism in P-glycoprotein.
Elife, 12, 2024
|
|
6DHI
 
 | |
5L79
 
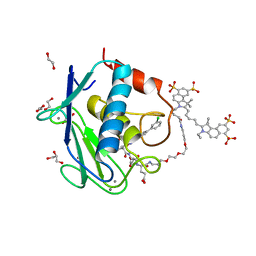 | | Crystal structure of MMP12 in complex with RXP470.1 conjugated with fluorophore Cy5,5 in space group P21212. | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CY5.5-PEG2, ... | | Authors: | Tepshi, L, Bordenave, T, Devel, L, Dive, V, Stura, E.A. | | Deposit date: | 2016-06-02 | | Release date: | 2016-09-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Synthesis and in Vitro and in Vivo Evaluation of MMP-12 Selective Optical Probes.
Bioconjug.Chem., 27, 2016
|
|
7ZKB
 
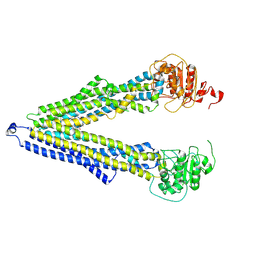 | | ABCB1 V978C mutant (mABCB1) in the inward facing state | | Descriptor: | (4S,11S,18S)-4-[[(2,4-dinitrophenyl)disulfanyl]methyl]-11,18-dimethyl-6,13,20-trithia-3,10,17,22,23,24-hexazatetracyclo[17.2.1.1^{5,8}.1^{12,15}]tetracosa-1(21),5(24),7,12(23),14,19(22)-hexaene-2,9,16-trione, (4~{S},11~{S},18~{S})-4,11-dimethyl-18-(sulfanylmethyl)-6,13,20-trithia-3,10,17,22,23,24-hexazatetracyclo[17.2.1.1^{5,8}.1^{12,15}]tetracosa-1(21),5(24),7,12(23),14,19(22)-hexaene-2,9,16-trione, ATP-dependent translocase ABCB1 | | Authors: | Parey, K, Januliene, D, Gewering, T, Moeller, A. | | Deposit date: | 2022-04-12 | | Release date: | 2023-04-26 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Tracing the substrate translocation mechanism in P-glycoprotein.
Elife, 12, 2024
|
|
7A9V
 
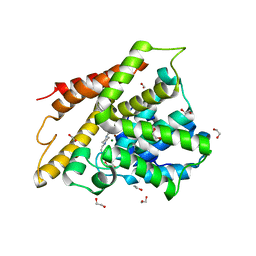 | | Crystal structure of human phosphodiesterase 4D2 catalytic domain with inhibitor NPD-635 | | Descriptor: | 1,2-ETHANEDIOL, 1-cycloheptyl-3-[4-methoxy-3-(2-phenylethoxy)phenyl]-4,4-dimethyl-4,5-dihydro-1H-pyrazol-5-one, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Singh, A.K, Blaazer, A.R, Zara, L, de Esch, I.J.P, Leurs, R, Brown, D.G. | | Deposit date: | 2020-09-02 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | hPDE4D2 structure with inhibitor NPD-635
To be published
|
|
5BUV
 
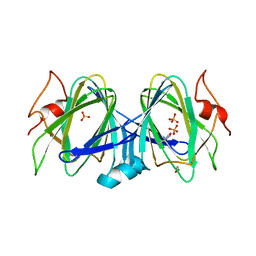 | | X-ray structure of WbcA from Yersinia enterocolitica | | Descriptor: | 1,2-ETHANEDIOL, 6-AMINOPYRIMIDIN-2(1H)-ONE, PHOSPHATE ION, ... | | Authors: | Holden, H.M, Salinger, A.J, Brown, H.A, Thoden, J.B. | | Deposit date: | 2015-06-04 | | Release date: | 2015-08-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Biochemical studies on WbcA, a sugar epimerase from Yersinia enterocolitica.
Protein Sci., 24, 2015
|
|
7AB9
 
 | | Crystal structure of human phosphodiesterase 4D2 catalytic domain with inhibitor NPD-656 | | Descriptor: | 1,2-ETHANEDIOL, 1-cycloheptyl-3-{4-methoxy-3-[2-(4-methoxyphenyl)ethoxy]phenyl}-4,4-dimethyl-4,5-dihydro-1H-pyrazol-5-one, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Singh, A.K, Blaazer, A.R, Zara, L, de Esch, I.J.P, Leurs, R, Brown, D.G. | | Deposit date: | 2020-09-07 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | hPDE4D2 structure with inhibitor NPD-656
To be published
|
|
6P8Y
 
 | |
6TGD
 
 | | Crystal structure of NDM-1 in complex with triazole-based inhibitor OP31 | | Descriptor: | 4-[[(2~{S})-oxolan-2-yl]methyl]-3-pyridin-3-yl-1~{H}-1,2,4-triazole-5-thione, CALCIUM ION, Metallo-beta-lactamase NDM-1, ... | | Authors: | Maso, L, Spirakis, F, Santucci, M, Simon, C, Docquier, J.D, Cruciani, G, Costi, M.P, Tondi, D, Cendron, L. | | Deposit date: | 2019-11-15 | | Release date: | 2020-08-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Virtual screening identifies broad-spectrum beta-lactamase inhibitors with activity on clinically relevant serine- and metallo-carbapenemases.
Sci Rep, 10, 2020
|
|
9BYJ
 
 | | Crystal Structure of Hck in complex with the Src-family kinase inhibitor A-419259 | | Descriptor: | 1,2-ETHANEDIOL, 7-[trans-4-(4-methylpiperazin-1-yl)cyclohexyl]-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine, DIMETHYL SULFOXIDE, ... | | Authors: | Selzer, A.M, Alvarado, J.J, Smithgall, T.E. | | Deposit date: | 2024-05-23 | | Release date: | 2024-10-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cocrystallization of the Src-Family Kinase Hck with the ATP-Site Inhibitor A-419259 Stabilizes an Extended Activation Loop Conformation.
Biochemistry, 63, 2024
|
|
3KHF
 
 | | The crystal structure of the PDZ domain of human Microtubule Associated Serine/Threonine Kinase 3 (MAST3) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Microtubule-associated serine/threonine-protein kinase 3 | | Authors: | Roos, A, Elkins, J, Savitsky, P, Wang, J, Ugochukwu, E, Murray, J, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A, von Delft, F, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-10-30 | | Release date: | 2009-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The crystal structure of the PDZ domain of human Microtubule Associated Serine/Threonine Kinase 3 (MAST3)
To be Published
|
|
6TD9
 
 | | X-ray structure of mature PA1624 from Pseudomonas aeruginosa PAO1 | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, PA1624, ... | | Authors: | Feiler, C.G, Blankenfeldt, W. | | Deposit date: | 2019-11-08 | | Release date: | 2019-12-04 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | The hypothetical periplasmic protein PA1624 from Pseudomonas aeruginosa folds into a unique two-domain structure.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
6GG8
 
 | | Mineralocorticoid receptor in complex with (s)-13 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Mineralocorticoid receptor, Nuclear receptor coactivator 1, ... | | Authors: | Edman, K, Aagaard, A, Tangefjord, S. | | Deposit date: | 2018-05-03 | | Release date: | 2019-01-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification of Mineralocorticoid Receptor Modulators with Low Impact on Electrolyte Homeostasis but Maintained Organ Protection.
J.Med.Chem., 62, 2019
|
|
6ZX2
 
 | | OMPD-domain of human UMPS in complex with 6-carboxamido-UMP at 1.2 Angstroms resolution | | Descriptor: | PROLINE, SULFATE ION, Uridine 5'-monophosphate synthase, ... | | Authors: | Tittmann, K, Rindfleisch, S, Schimdt, T. | | Deposit date: | 2020-07-29 | | Release date: | 2022-02-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Ground-state destabilization by electrostatic repulsion is not a driving force in orotidine-5-monophosphate decarboxylase catalysis
Nat Catal, 5, 2022
|
|
4Y0A
 
 | | Shikimate kinase from Acinetobacter baumannii in complex with shikimate | | Descriptor: | (3R,4S,5R)-3,4,5-TRIHYDROXYCYCLOHEX-1-ENE-1-CARBOXYLIC ACID, SULFATE ION, Shikimate kinase | | Authors: | Sutton, K.A, Breen, J, MacDonald, U, Beanan, J.M, Olson, R, Russo, T.A, Schultz, L.W, Umland, T.C. | | Deposit date: | 2015-02-05 | | Release date: | 2015-08-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.911 Å) | | Cite: | Structure of shikimate kinase, an in vivo essential metabolic enzyme in the nosocomial pathogen Acinetobacter baumannii, in complex with shikimate.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
6GIZ
 
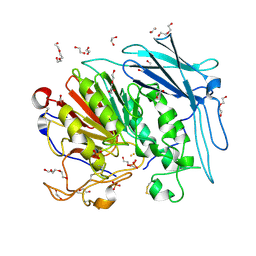 | | PURPLE ACID PHYTASE FROM WHEAT ISOFORM B2 - SUBSTRATE COMPLEX | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Faba-Rodriguez, R, Brearley, C.A, Hemmings, A.M. | | Deposit date: | 2018-05-15 | | Release date: | 2019-11-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structure of a cereal purple acid phytase provides new insights to phytate degradation in plants.
Plant Commun., 3, 2022
|
|
9HGB
 
 | | Amyloid fibril from the antimicrobial peptide citropin 1-3 - Polymorph Nr.3 | | Descriptor: | Citropin-1.3 | | Authors: | Strati, F, Rayan, B, Landau, M, Siavash, M, Cali, P.M, Monistrol, J, Golubev, A, Gustavsson, E, Bloch, Y. | | Deposit date: | 2024-11-19 | | Release date: | 2025-10-01 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of citropin 1.3 amyloid polymorph
To Be Published
|
|
4Y2U
 
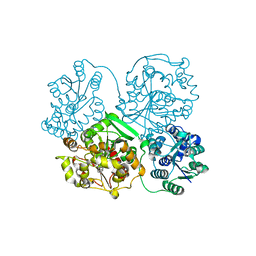 | | Structure of soluble epoxide hydrolase in complex with tert-butyl 1,2,3,4-tetrahydroquinolin-3-ylcarbamate | | Descriptor: | Bifunctional epoxide hydrolase 2, MAGNESIUM ION, tert-butyl (3R)-1,2,3,4-tetrahydroquinolin-3-ylcarbamate | | Authors: | Amano, Y, Yamaguchi, T. | | Deposit date: | 2015-02-10 | | Release date: | 2015-05-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Identification of N-ethylmethylamine as a novel scaffold for inhibitors of soluble epoxide hydrolase by crystallographic fragment screening
Bioorg.Med.Chem., 23, 2015
|
|
4HJH
 
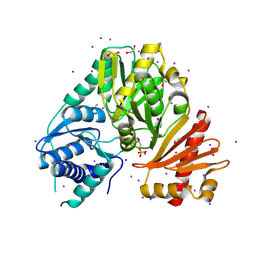 | |
6PNL
 
 | | Structure of Epimerase Mth375 from the thermophilic pseudomurein-containing methanogen Methanothermobacter thermautotrophicus | | Descriptor: | 1,2-ETHANEDIOL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Carbone, V, Schofield, L.R, Sutherland-Smith, A.J, Ronimus, R.S. | | Deposit date: | 2019-07-02 | | Release date: | 2020-07-08 | | Last modified: | 2025-09-17 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural characterisation of nucleotide sugar short-chain dehydrogenases/reductases from the thermophilic pseudomurein-containing methanogen Methanothermobacter thermautotrophicus Delta H.
Febs J., 2025
|
|
6GJ2
 
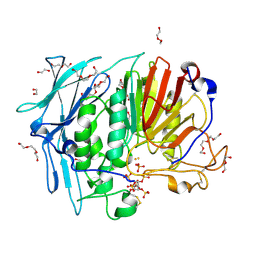 | | PURPLE ACID PHYTASE FROM WHEAT ISOFORM B2 - COMPLEX WITH INOSITOL HEXASULPHATE | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Faba-Rodriguez, R, Brearley, C.A, Hemmings, A.M. | | Deposit date: | 2018-05-15 | | Release date: | 2019-11-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structure of a cereal purple acid phytase provides new insights to phytate degradation in plants.
Plant Commun., 3, 2022
|
|
5B21
 
 | |
8AAA
 
 | | Crystal structure of SARS-CoV-2 S RBD in complex with a stapled peptide | | Descriptor: | 1,1',1''-(1,3,5-triazinane-1,3,5-triyl)tripropan-1-one, Spike protein S1, Stapled peptide | | Authors: | Brear, P, Chen, L, Gaynor, K, Harman, M, Dods, R, Hyvonen, M. | | Deposit date: | 2022-06-30 | | Release date: | 2023-06-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Multivalent bicyclic peptides are an effective antiviral modality that can potently inhibit SARS-CoV-2.
Nat Commun, 14, 2023
|
|
