4MA5
 
 | | The crystal structure of phosphoribosylaminoimidazole carboxylase ATPase subunit of Francisella tularensis subsp. tularensis SCHU S4 in complex with an ATP analog, AMP-PNP. | | Descriptor: | FORMIC ACID, GLYCEROL, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Tan, K, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-08-15 | | Release date: | 2013-08-28 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.809 Å) | | Cite: | The crystal structure of phosphoribosylaminoimidazole carboxylase ATPase subunit of Francisella tularensis subsp. tularensis SCHU S4 in complex with an ATP analog, AMP-PNP.
To be Published
|
|
4M9D
 
 | | The Crystal structure of an adenylosuccinate synthetase from Bacillus anthracis str. Ames Ancestor in complex with AMP. | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE MONOPHOSPHATE, Adenylosuccinate synthetase, ... | | Authors: | Tan, K, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-08-14 | | Release date: | 2013-08-28 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.821 Å) | | Cite: | The Crystal structure of an adenylosuccinate synthetase from Bacillus anthracis str. Ames Ancestor in complex with AMP.
To be Published
|
|
7BES
 
 | | CryoEM structure of Mycobacterium tuberculosis UMP Kinase (UMPK) in complex with UDP and UTP | | Descriptor: | URIDINE 5'-TRIPHOSPHATE, URIDINE-5'-DIPHOSPHATE, Uridylate kinase | | Authors: | Bous, J, Trapani, S, Walter, P, Bron, P, Munier-Lehmann, H. | | Deposit date: | 2020-12-24 | | Release date: | 2022-01-12 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Structural basis for the allosteric inhibition of UMP kinase from Gram-positive bacteria, a promising antibacterial target.
Febs J., 289, 2022
|
|
5CLF
 
 | |
4JDB
 
 | | Structure of Trypanosoma cruzi dihydroorotate dehydrogenase in complex with MII-5-005 | | Descriptor: | 5-[2-(7-methoxynaphthalen-2-yl)ethyl]-2,6-dioxo-1,2,3,6-tetrahydropyrimidine-4-carboxylic acid, COBALT HEXAMMINE(III), Dihydroorotate dehydrogenase (fumarate), ... | | Authors: | Inaoka, D.K, Iida, M, Tabuchi, T, Lee, N, Hashimoto, S, Matsuoka, S, Kuranaga, T, Shiba, T, Sakamoto, K, Suzuki, S, Balogun, E.O, Nara, T, Aoki, T, Inoue, M, Honma, T, Tanaka, A, Harada, S, Kita, K. | | Deposit date: | 2013-02-24 | | Release date: | 2014-02-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structure of Trypanosoma cruzi dihydroorotate dehydrogenase in complex with MII-5-005
To be Published
|
|
7L0K
 
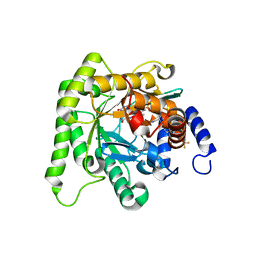 | | Crystal structure of Plasmodium falciparum dihydroorotate dehydrogenase bound with Inhibitor DSM784 (3-(1-(3-methyl-4-((6-(trifluoromethyl)pyridin-3-yl)methyl)-1H-pyrrole-2-carboxamido)ethyl)-1H-pyrazole-5-carboxamide) | | Descriptor: | 3-{(1R)-1-[(3-methyl-4-{[6-(trifluoromethyl)pyridin-3-yl]methyl}-1H-pyrrole-2-carbonyl)amino]ethyl}-1H-pyrazole-5-carboxamide, Dihydroorotate dehydrogenase (quinone), mitochondrial, ... | | Authors: | Deng, X, Phillips, M, Tomchick, D. | | Deposit date: | 2020-12-11 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Potent Antimalarials with Development Potential Identified by Structure-Guided Computational Optimization of a Pyrrole-Based Dihydroorotate Dehydrogenase Inhibitor Series.
J.Med.Chem., 64, 2021
|
|
7KZY
 
 | | Crystal structure of Plasmodium falciparum dihydroorotate dehydrogenase bound with Inhibitor DSM778 (3-methyl-N-(1-(5-methyl-1H-pyrazol-3-yl)ethyl)-4-(1-(6-(trifluoromethyl)pyridin-3-yl)cyclopropyl)-1H-pyrrole-2-carboxamide) | | Descriptor: | 3-methyl-N-[(1R)-1-(5-methyl-1H-pyrazol-3-yl)ethyl]-4-{1-[6-(trifluoromethyl)pyridin-3-yl]cyclopropyl}-1H-pyrrole-2-carboxamide, Dihydroorotate dehydrogenase (quinone), mitochondrial, ... | | Authors: | Deng, X, Phillips, M, Tomchick, D. | | Deposit date: | 2020-12-10 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Potent Antimalarials with Development Potential Identified by Structure-Guided Computational Optimization of a Pyrrole-Based Dihydroorotate Dehydrogenase Inhibitor Series.
J.Med.Chem., 64, 2021
|
|
7KYK
 
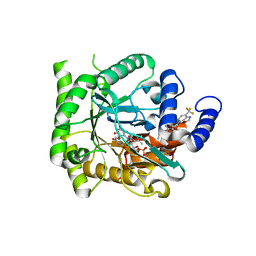 | | Crystal structure of Plasmodium falciparum dihydroorotate dehydrogenase bound with Inhibitor DSM589 (ethyl 3-methyl-4-((4-(trifluoromethyl)benzo[d]oxazol-7-yl)methyl)-1H-pyrrole-2-carboxylate) | | Descriptor: | Dihydroorotate dehydrogenase (quinone), mitochondrial, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Deng, X, Phillips, M, Tomchick, D. | | Deposit date: | 2020-12-08 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Potent Antimalarials with Development Potential Identified by Structure-Guided Computational Optimization of a Pyrrole-Based Dihydroorotate Dehydrogenase Inhibitor Series.
J.Med.Chem., 64, 2021
|
|
7KZ4
 
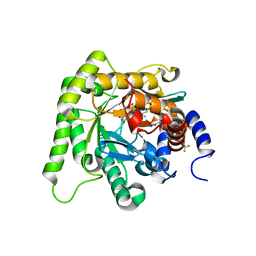 | | Crystal structure of Plasmodium falciparum dihydroorotate dehydrogenase bound with Inhibitor DSM705 (N-(1-(1H-1,2,4-triazol-3-yl)ethyl)-3-methyl-4-(1-(6-(trifluoromethyl)pyridin-3-yl)cyclopropyl)-1H-pyrrole-2-carboxamide) | | Descriptor: | 3-methyl-N-[(1R)-1-(1H-1,2,4-triazol-3-yl)ethyl]-4-{1-[6-(trifluoromethyl)pyridin-3-yl]cyclopropyl}-1H-pyrrole-2-carboxamide, Dihydroorotate dehydrogenase (quinone), mitochondrial, ... | | Authors: | Deng, X, Phillips, M, Tomchick, D. | | Deposit date: | 2020-12-09 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Potent Antimalarials with Development Potential Identified by Structure-Guided Computational Optimization of a Pyrrole-Based Dihydroorotate Dehydrogenase Inhibitor Series.
J.Med.Chem., 64, 2021
|
|
7KYY
 
 | | Crystal structure of Plasmodium falciparum dihydroorotate dehydrogenase bound with Inhibitor DSM697 (3-methyl-N-(1-(5-methylisoxazol-3-yl)ethyl)-4-(6-(trifluoromethyl)-1H-indol-3-yl)-1H-pyrrole-2-carboxamide) | | Descriptor: | 3-methyl-N-[(1R)-1-(5-methyl-1,2-oxazol-3-yl)ethyl]-4-[6-(trifluoromethyl)-1H-indol-3-yl]-1H-pyrrole-2-carboxamide, Dihydroorotate dehydrogenase (quinone), mitochondrial, ... | | Authors: | Deng, X, Phillips, M, Tomchick, D. | | Deposit date: | 2020-12-09 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Potent Antimalarials with Development Potential Identified by Structure-Guided Computational Optimization of a Pyrrole-Based Dihydroorotate Dehydrogenase Inhibitor Series.
J.Med.Chem., 64, 2021
|
|
7L01
 
 | | Crystal structure of Plasmodium falciparum dihydroorotate dehydrogenase bound with Inhibitor DSM782 (N-(1-(5-cyano-1H-pyrazol-3-yl)ethyl)-3-methyl-4-(1-(6-(trifluoromethyl)pyridin-3-yl)cyclopropyl)-1H-pyrrole-2-carboxamide) | | Descriptor: | Dihydroorotate dehydrogenase (quinone), mitochondrial, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Deng, X, Phillips, M, Tomchick, D. | | Deposit date: | 2020-12-10 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Potent Antimalarials with Development Potential Identified by Structure-Guided Computational Optimization of a Pyrrole-Based Dihydroorotate Dehydrogenase Inhibitor Series.
J.Med.Chem., 64, 2021
|
|
7KYV
 
 | | Crystal structure of Plasmodium falciparum dihydroorotate dehydrogenase bound with Inhibitor DSM634 (3-methyl-N-(1-(5-methylisoxazol-3-yl)ethyl)-4-(4-(trifluoromethyl)benzyl)-1H-pyrrole-2-carboxamide) | | Descriptor: | 3-methyl-N-[(1R)-1-(5-methyl-1,2-oxazol-3-yl)ethyl]-4-{[4-(trifluoromethyl)phenyl]methyl}-1H-pyrrole-2-carboxamide, Dihydroorotate dehydrogenase (quinone), mitochondrial, ... | | Authors: | Deng, X, Phillips, M, Tomchick, D. | | Deposit date: | 2020-12-08 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Potent Antimalarials with Development Potential Identified by Structure-Guided Computational Optimization of a Pyrrole-Based Dihydroorotate Dehydrogenase Inhibitor Series.
J.Med.Chem., 64, 2021
|
|
3AV3
 
 | | Crystal structure of glycinamide ribonucleotide transformylase 1 from Geobacillus kaustophilus | | Descriptor: | MAGNESIUM ION, Phosphoribosylglycinamide formyltransferase | | Authors: | Kanagawa, M, Baba, S, Nakagawa, N, Ebihara, A, Kuramitsu, S, Yokoyama, S, Sampei, G, Kawai, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2011-02-18 | | Release date: | 2012-03-07 | | Last modified: | 2014-01-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures and reaction mechanisms of the two related enzymes, PurN and PurU.
J.Biochem., 154, 2013
|
|
5CLI
 
 | |
5CLJ
 
 | |
2R00
 
 | | crystal structure of aspartate semialdehyde dehydrogenase II complexed with ASA from vibrio cholerae | | Descriptor: | 2,2'-oxydiacetic acid, Aspartate-semialdehyde dehydrogenase | | Authors: | Viola, R.E, Liu, X, Ohren, J.F, Faehnle, C.R. | | Deposit date: | 2007-08-17 | | Release date: | 2008-04-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | The structure of a redundant enzyme: a second isoform of aspartate beta-semialdehyde dehydrogenase in Vibrio cholerae.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
3AUF
 
 | | Crystal structure of glycinamide ribonucleotide transformylase 1 from Symbiobacterium toebii | | Descriptor: | Glycinamide ribonucleotide transformylase 1 | | Authors: | Kanagawa, M, Baba, S, Nagira, T, Kuramitsu, S, Yokoyama, S, Sampei, G, Kawai, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2011-02-03 | | Release date: | 2012-03-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structures and reaction mechanisms of the two related enzymes, PurN and PurU.
J.Biochem., 154, 2013
|
|
2GYY
 
 | |
3AW8
 
 | | Crystal structure of N5-carboxyaminoimidazole ribonucleotide synthetase from Thermus thermophilus HB8 | | Descriptor: | ADENOSINE MONOPHOSPHATE, CHLORIDE ION, Phosphoribosylaminoimidazole carboxylase, ... | | Authors: | Okada, K, Tsunoda, S, Taka, H, Baba, S, Kanagawa, M, Nakagawa, N, Ebihara, A, Kuramitsu, S, Yokoyama, S, Kawai, G, Sampei, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2011-03-15 | | Release date: | 2012-04-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of N5-carboxyaminoimidazole ribonucleotide synthetase, PurK, from thermophilic bacteria
To be Published
|
|
2GZ1
 
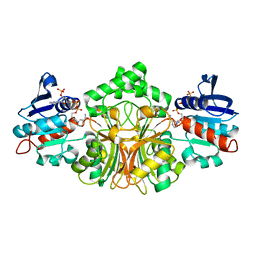 | | Structure of Aspartate Semialdehyde Dehydrogenase (ASADH) from Streptococcus pneumoniae complexed with NADP | | Descriptor: | Aspartate beta-semialdehyde dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Faehnle, C.R, Le Coq, J, Liu, X, Viola, R.E. | | Deposit date: | 2006-05-10 | | Release date: | 2006-08-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Examination of key intermediates in the catalytic cycle of aspartate-beta-semialdehyde dehydrogenase from a gram-positive infectious bacteria.
J.Biol.Chem., 281, 2006
|
|
2VHH
 
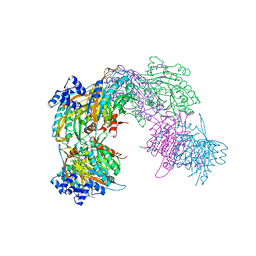 | | Crystal structure of a pyrimidine degrading enzyme from Drosophila melanogaster | | Descriptor: | CG3027-PA | | Authors: | Lundgren, S, Lohkamp, B, Andersen, B, Piskur, J, Dobritzsch, D. | | Deposit date: | 2007-11-21 | | Release date: | 2008-03-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Crystal Structure of Beta-Alanine Synthase from Drosophila Melanogaster Reveals a Homooctameric Helical Turn-Like Assembly.
J.Mol.Biol., 377, 2008
|
|
2D0T
 
 | | Crystal structure of 4-phenylimidazole bound form of human indoleamine 2,3-dioxygenase | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, 4-PHENYL-1H-IMIDAZOLE, Indoleamine 2,3-dioxygenase, ... | | Authors: | Sugimoto, H, Oda, S, Otsuki, T, Hino, T, Yoshida, T, Shiro, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-08-08 | | Release date: | 2006-01-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of human indoleamine 2,3-dioxygenase: catalytic mechanism of O2 incorporation by a heme-containing dioxygenase.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
5CVT
 
 | | Structure of a single tryptophan mutant of Acetobacter aceti PurE containing 5-fluorotryptophan, pH 5.4 | | Descriptor: | 2,5,8,11-TETRAOXATRIDECANE, ACETATE ION, CITRIC ACID, ... | | Authors: | Sullivan, K.L, Kappock, T.J. | | Deposit date: | 2015-07-27 | | Release date: | 2016-08-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structure of a single tryptophan mutant of Acetobacter aceti PurE
To Be Published
|
|
2PPO
 
 | |
5DEL
 
 | |
