7CD9
 
 | | Crystal Structure of SETDB1 tudor domain in complexed with Compound 6 | | Descriptor: | 3-methyl-2-[[(3R,5R)-1-methyl-5-(4-phenylmethoxyphenyl)piperidin-3-yl]amino]-5H-pyrrolo[3,2-d]pyrimidin-4-one, CITRIC ACID, Histone-lysine N-methyltransferase SETDB1 | | Authors: | Xiong, L, Guo, Y, Mao, X, Huang, L, Wu, C, Yang, S. | | Deposit date: | 2020-06-19 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-Guided Discovery of a Potent and Selective Cell-Active Inhibitor of SETDB1 Tudor Domain.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7V1U
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with the inhibitor ZJ12 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, ~{N}-(3-ethyl-6-methoxy-1,2-benzoxazol-5-yl)-2-methoxy-benzenesulfonamide | | Authors: | Zhang, M, Wang, C, Zhang, C, Zhang, Y, Xu, Y. | | Deposit date: | 2021-08-06 | | Release date: | 2022-08-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Design, synthesis and pharmacological characterization of N-(3-ethylbenzo[d]isoxazol-5-yl) sulfonamide derivatives as BRD4 inhibitors against acute myeloid leukemia.
Acta Pharmacol.Sin., 43, 2022
|
|
7KYE
 
 | | Structure of a GNAT superfamily PA3944 acetyltransferase in complex with CHES | | Descriptor: | 1,2-ETHANEDIOL, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Acetyltransferase PA3944, ... | | Authors: | Czub, M.P, Porebski, P.J, Cymborowski, M, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-12-07 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structure of a GNAT superfamily PA3944 acetyltransferase in complex with CHES
To Be Published
|
|
9I4H
 
 | | Factor Inhibiting HIF (FIH) in complex with manganese and 3-Hydroxy-5-(3-(4-(hydroxymethyl)-3-nitrophenyl)isoxazol-5-yl)picolinoyl)glycine | | Descriptor: | 2-[[5-[3-[4-(hydroxymethyl)-3-nitro-phenyl]-1,2-oxazol-5-yl]-3-oxidanyl-pyridin-2-yl]carbonylamino]ethanoic acid, Hypoxia-inducible factor 1-alpha inhibitor, MANGANESE (II) ION | | Authors: | Kaur, S, Zhang, X.J, Schofield, C.J. | | Deposit date: | 2025-01-24 | | Release date: | 2025-05-21 | | Last modified: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Light-Induced, Lysine-Targeting Irreversible Covalent Inhibition of the Human Oxygen Sensing Hydroxylase Factor Inhibiting HIF (FIH).
J.Am.Chem.Soc., 147, 2025
|
|
5V5I
 
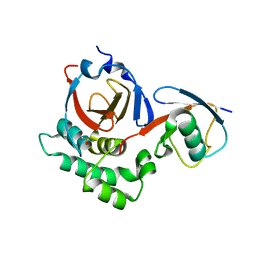 | |
5HVM
 
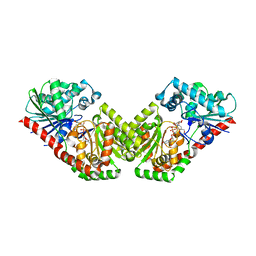 | | Structure of Aspergillus fumigatus trehalose-6-phosphate synthase A in complex with UDP and validoxylamine A | | Descriptor: | (1S,2S,3R,6S)-4-(HYDROXYMETHYL)-6-{[(1S,2S,3S,4R,5R)-2,3,4-TRIHYDROXY-5-(HYDROXYMETHYL)CYCLOHEXYL]AMINO}CYCLOHEX-4-ENE-1,2,3-TRIOL, Alpha,alpha-trehalose-phosphate synthase (UDP-forming), URIDINE-5'-DIPHOSPHATE | | Authors: | Miao, Y, Brennan, R.G. | | Deposit date: | 2016-01-28 | | Release date: | 2017-05-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.815 Å) | | Cite: | Structural and In Vivo Studies on Trehalose-6-Phosphate Synthase from Pathogenic Fungi Provide Insights into Its Catalytic Mechanism, Biological Necessity, and Potential for Novel Antifungal Drug Design.
MBio, 8, 2017
|
|
6DOG
 
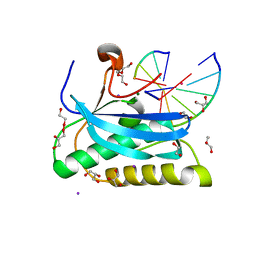 | |
6DPN
 
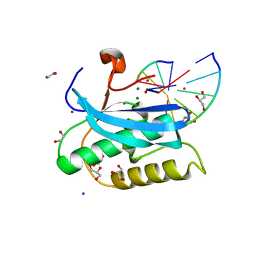 | |
6CJH
 
 | | Co-crystal structure of MNK2 in complex with an inhibitor | | Descriptor: | 3-phenyl-5-(pyridin-4-yl)-1H-indazole, MAP kinase-interacting serine/threonine-protein kinase 2, ZINC ION | | Authors: | Han, Q. | | Deposit date: | 2018-02-26 | | Release date: | 2018-05-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structure-based Design of Pyridone-Aminal eFT508 Targeting Dysregulated Translation by Selective Mitogen-activated Protein Kinase Interacting Kinases 1 and 2 (MNK1/2) Inhibition.
J. Med. Chem., 61, 2018
|
|
6DPD
 
 | |
7KPP
 
 | | Structure of the E102A mutant of a GNAT superfamily PA3944 acetyltransferase | | Descriptor: | 1,2-ETHANEDIOL, Acetyltransferase PA3944, COENZYME A, ... | | Authors: | Czub, M.P, Porebski, P.J, Majorek, K.A, Cymborowski, M, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-11-12 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Gcn5-Related N- Acetyltransferases (GNATs) With a Catalytic Serine Residue Can Play Ping-Pong Too.
Front Mol Biosci, 8, 2021
|
|
2V1X
 
 | | Crystal structure of human RECQ-like DNA helicase | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, ATP-DEPENDENT DNA HELICASE Q1, ... | | Authors: | Pike, A.C.W, Shrestha, B, Burgess-Brown, N, King, O, Ugochukwu, E, Watt, S, Edwards, A, Arrowsmith, C.H, Weigelt, J, Sundstrom, M, Gileadi, O. | | Deposit date: | 2007-05-30 | | Release date: | 2007-07-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the Human Recq1 Helicase Reveals a Putative Strand-Separation Pin.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
7BZJ
 
 | | The Discovery of Benzhydrol-Oxaborole Hybrid Derivatives as Leucyl-tRNA Synthetase Inhibitors | | Descriptor: | Leucine--tRNA ligase, [(1~{R},5~{R},6~{S},8~{R})-8-(6-aminopurin-9-yl)-4'-[(~{R})-oxidanyl-[4-(2-oxidanylidenepropylsulfanyl)phenyl]methyl]spiro[2,4,7-trioxa-3-boranuidabicyclo[3.3.0]octane-3,7'-7-boranuidabicyclo[4.3.0]nona-1(6),2,4-triene]-6-yl]methoxy-tris(oxidanyl)phosphanium | | Authors: | Liu, R.J, Li, H, Wang, E.D, Zhou, H. | | Deposit date: | 2020-04-28 | | Release date: | 2020-12-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of benzhydrol-oxaborole derivatives as Streptococcus pneumoniae leucyl-tRNA synthetase inhibitors.
Bioorg.Med.Chem., 29, 2021
|
|
3ZXJ
 
 | | Engineering the active site of a GH43 glycoside hydrolase generates a biotechnologically significant enzyme that displays both endo- xylanase and exo-arabinofuranosidase activity | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, HIAXHD3, ... | | Authors: | McKee, L.S, Pena, M.J, Rogowski, A, Jackson, A, Lewis, R.J, York, W.S, Krogh, K.B.R.M, Vikso-Nielsen, A, Skjot, M, Gilbert, H.J, Marles-Wright, J. | | Deposit date: | 2011-08-11 | | Release date: | 2012-04-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Introducing Endo-Xylanase Activity Into an Exo-Acting Arabinofuranosidase that Targets Side Chains.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3M3M
 
 | | Crystal structure of glutathione S-transferase from Pseudomonas fluorescens [Pf-5] | | Descriptor: | 1,2-ETHANEDIOL, GLUTATHIONE, Glutathione S-transferase, ... | | Authors: | Bagaria, A, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-03-09 | | Release date: | 2010-03-16 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of glutathione S-transferase from Pseudomonas fluorescens [Pf-5]
To be Published
|
|
5BX2
 
 | | Crystal structure of Thermoanaerobacterium xylanolyticum GH116 beta-glucosidase with 2-deoxy-2-fluoroglucoside | | Descriptor: | 1,2-ETHANEDIOL, 2-deoxy-2-fluoro-alpha-D-glucopyranose, CALCIUM ION, ... | | Authors: | Charoenwattanasatien, R, Pengthaisong, S, Sansenya, S, Mutoh, R, Tanaka, H, Kurisu, G, Ketudat Cairns, J.R. | | Deposit date: | 2015-06-08 | | Release date: | 2016-05-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Bacterial beta-Glucosidase Reveals the Structural and Functional Basis of Genetic Defects in Human Glucocerebrosidase 2 (GBA2)
Acs Chem.Biol., 11, 2016
|
|
5HNX
 
 | | Structural basis of backwards motion in kinesin-14: minus-end directed nKn664 in the nucleotide-free state | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Shigematsu, H, Yokoyama, T, Kikkawa, M, Shirouzu, M, Nitta, R. | | Deposit date: | 2016-01-19 | | Release date: | 2016-08-10 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Structural Basis of Backwards Motion in Kinesin-1-Kinesin-14 Chimera: Implication for Kinesin-14 Motility
Structure, 24, 2016
|
|
1UU2
 
 | | Histidinol-phosphate aminotransferase (HisC) from Thermotoga maritima (apo-form) | | Descriptor: | 1,2-ETHANEDIOL, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, HISTIDINOL-PHOSPHATE AMINOTRANSFERASE | | Authors: | Vega, M.C, Fernandez, F.J, Lehmann, F, Wilmanns, M. | | Deposit date: | 2003-12-13 | | Release date: | 2004-03-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Studies of the Catalytic Reaction Pathway of a Hyperthermophilic Histidinol-Phosphate Aminotransferase
J.Biol.Chem., 279, 2004
|
|
5ZS7
 
 | |
5C05
 
 | | Crystal Structure of Gamma-terpinene Synthase from Thymus vulgaris | | Descriptor: | 1,2-ETHANEDIOL, Putative gamma-terpinene synthase | | Authors: | Parthier, C.P, Rudolph, K, Muller, Y.A, Mueller-Uri, F. | | Deposit date: | 2015-06-12 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Expression, crystallization and structure elucidation of gamma-terpinene synthase from Thymus vulgaris.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
6DOB
 
 | |
6DOL
 
 | |
6IX5
 
 | | The structure of LepI complex with SAM and its substrate analogue | | Descriptor: | 1,2-ETHANEDIOL, 4-hydroxy-3-[(2S,6E,8E)-2-methyldeca-6,8-dienoyl]-5-phenylpyridin-2(1H)-one, CHLORIDE ION, ... | | Authors: | Cai, Y, Ohashi, M, Hai, Y, Tang, Y, Zhou, J. | | Deposit date: | 2018-12-09 | | Release date: | 2019-07-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for stereoselective dehydration and hydrogen-bonding catalysis by the SAM-dependent pericyclase LepI.
Nat.Chem., 11, 2019
|
|
6DPE
 
 | |
6G2E
 
 | | X-ray structure of NSD3-PWWP1 in complex with compound 13 | | Descriptor: | Histone-lysine N-methyltransferase NSD3, [3,5-dimethyl-4-(1-methyl-5-pyridin-4-yl-imidazol-4-yl)phenyl]methanamine | | Authors: | Boettcher, J, Muellauer, B.J, Weiss-Puxbaum, A, Zoephel, A. | | Deposit date: | 2018-03-23 | | Release date: | 2019-06-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Fragment-based discovery of a chemical probe for the PWWP1 domain of NSD3.
Nat.Chem.Biol., 15, 2019
|
|
