7T31
 
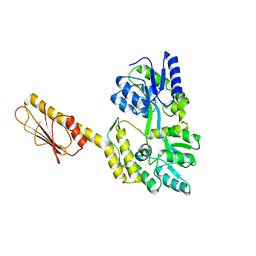 | | X-ray Structure of Clostridiodies difficile PilW | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Putative pilin protein chimera | | Authors: | Ronish, L.A, Piepenbrink, K.H. | | Deposit date: | 2021-12-06 | | Release date: | 2022-10-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Recognition of extracellular DNA by type IV pili promotes biofilm formation by Clostridioides difficile.
J.Biol.Chem., 298, 2022
|
|
3RKP
 
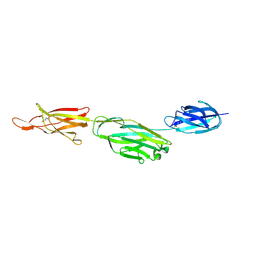 | | Crystal structure of BcpA*(D312A), the major pilin subunit of Bacillus cereus | | Descriptor: | Collagen adhesion protein | | Authors: | Hendrickx, A.P, Poor, C.B, Jureller, J.E, Budzik, J.M, He, C, Schneewind, O. | | Deposit date: | 2011-04-18 | | Release date: | 2012-05-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.243 Å) | | Cite: | Isopeptide bonds of the major pilin protein BcpA influence pilus structure and bundle formation on the surface of Bacillus cereus.
Mol.Microbiol., 85, 2012
|
|
5FAA
 
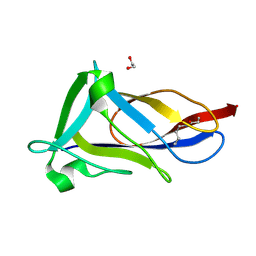 | | Crystal structure of C-terminal domain of shaft pilin spaA from Lactobacillus rhamnosus GG, - I422 space group | | Descriptor: | 1,2-ETHANEDIOL, Cell surface protein SpaA | | Authors: | Chaurasia, P, Pratap, S, von Ossowski, I, Palva, A, Krishnan, V. | | Deposit date: | 2015-12-11 | | Release date: | 2016-07-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | New insights about pilus formation in gut-adapted Lactobacillus rhamnosus GG from the crystal structure of the SpaA backbone-pilin subunit
Sci Rep, 6, 2016
|
|
4QS4
 
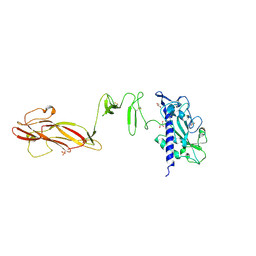 | |
6GMS
 
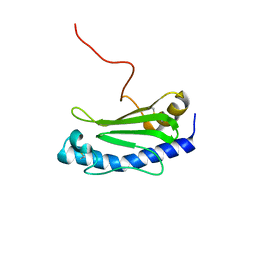 | | Solution NMR structure of the major type IV pilin PpdD from enterohemorrhagic Escherichia coli (EHEC) | | Descriptor: | Prepilin peptidase-dependent protein D | | Authors: | Amorim, G.C, Bardiaux, B, Luna-Rico, A, Zeng, W, Guilvout, I, Egelman, E, Nilges, M, Francetic, O, Izadi-Pruneyre, N. | | Deposit date: | 2018-05-28 | | Release date: | 2019-05-15 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure and Assembly of the Enterohemorrhagic Escherichia coli Type 4 Pilus.
Structure, 27, 2019
|
|
3JVU
 
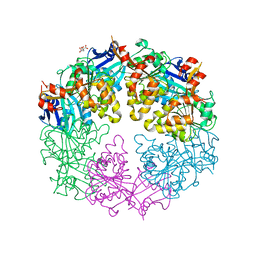 | |
4HSS
 
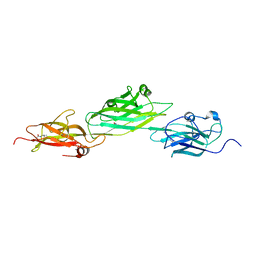 | |
4HSQ
 
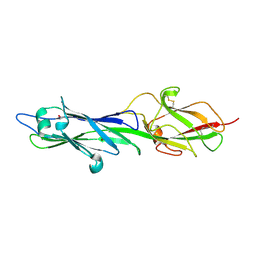 | |
2M5G
 
 | | Solution structure of FimA wt | | Descriptor: | Type-1 fimbrial protein, A chain | | Authors: | Walczak, M.J, Puorger, C, Glockshuber, R, Wider, G. | | Deposit date: | 2013-02-24 | | Release date: | 2013-11-13 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Intramolecular donor strand complementation in the E. coli type 1 pilus subunit FimA explains the existence of FimA monomers as off-pathway products of pilus assembly that inhibit host cell apoptosis.
J.Mol.Biol., 426, 2014
|
|
3RBK
 
 | |
2OPE
 
 | | Crystal structure of the Neisseria meningitidis minor Type IV pilin, PilX, in space group P43 | | Descriptor: | PilX | | Authors: | Dyer, D.H, Helaine, S, Pelicic, V, Forest, K.T. | | Deposit date: | 2007-01-29 | | Release date: | 2007-10-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 3D structure/function analysis of PilX reveals how minor pilins can modulate the virulence properties of type IV pili.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2OPD
 
 | | Structure of the Neisseria meningitidis minor Type IV pilin, PilX | | Descriptor: | PilX | | Authors: | Dyer, D.H, Helaine, S, Pelicic, V, Forest, K.T. | | Deposit date: | 2007-01-29 | | Release date: | 2007-10-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | 3D structure/function analysis of PilX reveals how minor pilins can modulate the virulence properties of type IV pili.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
1QPP
 
 | |
1QPX
 
 | |
6FJY
 
 | | Crystal structure of CsuC-CsuE chaperone-tip adhesion subunit pre-assembly complex from archaic chaperone-usher Csu pili of Acinetobacter baumannii | | Descriptor: | CsuC, Protein CsuE | | Authors: | Pakharukova, N.A, Tuitilla, M, Paavilainen, S, Zavialov, A.V. | | Deposit date: | 2018-01-23 | | Release date: | 2018-05-16 | | Last modified: | 2018-09-19 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structural basis forAcinetobacter baumanniibiofilm formation.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
3RBI
 
 | |
3RBJ
 
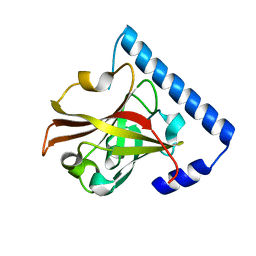 | |
8PJP
 
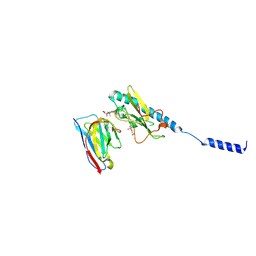 | | Neisseria meningitidis PilE, SB-GATDH variant, bound to the F10 nanobody | | Descriptor: | (2~{R})-~{N}-[(2~{R},3~{S},4~{S},5~{R},6~{R})-5-acetamido-2-methyl-4,6-bis(oxidanyl)oxan-3-yl]-2,3-bis(oxidanyl)propanamide, Nanobody F10, Pilin, ... | | Authors: | Fernandez-Martinez, D, Dumenil, G. | | Deposit date: | 2023-06-23 | | Release date: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Cryo-EM structures of type IV pili complexed with nanobodies reveal immune escape mechanisms.
Nat Commun, 15, 2024
|
|
3G66
 
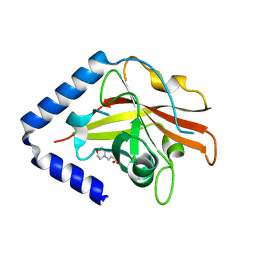 | | The crystal structure of Streptococcus pneumoniae Sortase C provides novel insights into catalysis as well as pilin substrate specificity | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Sortase C | | Authors: | Neiers, F, Madhurantakam, C, Falker, S, Manzano, C, Dessen, A, Normark, S, Henriques-Normark, B, Achour, A. | | Deposit date: | 2009-02-06 | | Release date: | 2009-09-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Two crystal structures of pneumococcal pilus sortase C provide novel insights into catalysis and substrate specificity.
J.Mol.Biol., 393, 2009
|
|
3B2M
 
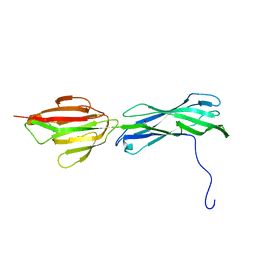 | |
3G69
 
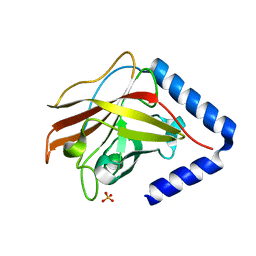 | | The crystal structure of Streptococcus pneumoniae Sortase C provides novel insights into catalysis as well as pilin substrate specificity | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, SULFATE ION, Sortase C | | Authors: | Neiers, F, Madhurantakam, C, Falker, S, Manzano, C, Dessen, A, Normark, S, Henriques-Normark, B, Achour, A. | | Deposit date: | 2009-02-06 | | Release date: | 2009-09-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Two crystal structures of pneumococcal pilus sortase C provide novel insights into catalysis and substrate specificity.
J.Mol.Biol., 393, 2009
|
|
5CYL
 
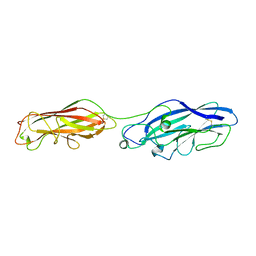 | |
5LP9
 
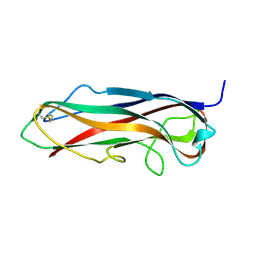 | | FimA wt from S. flexneri | | Descriptor: | Major type 1 subunit fimbrin (Pilin) | | Authors: | Zyla, D, Capitani, G, Prota, A, Glockshuber, R. | | Deposit date: | 2016-08-12 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (0.88626635 Å) | | Cite: | Alternative folding to a monomer or homopolymer is a common feature of the type 1 pilus subunit FimA from enteroinvasive bacteria.
J.Biol.Chem., 2019
|
|
8TUX
 
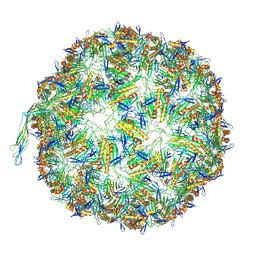 | |
3FKQ
 
 | |
