1GFC
 
 | |
1G2B
 
 | | ALPHA-SPECTRIN SRC HOMOLOGY 3 DOMAIN, CIRCULAR PERMUTANT, CUT AT N47-D48 | | Descriptor: | SPECTRIN ALPHA CHAIN, SULFATE ION | | Authors: | Berisio, R, Viguera, A.R, Serrano, L, Wilmanns, M. | | Deposit date: | 2000-10-18 | | Release date: | 2000-11-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Atomic resolution structure of a mutant of the spectrin SH3 domain.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1HD3
 
 | | A-SPECTRIN SH3 DOMAIN F52Y MUTANT | | Descriptor: | GLYCEROL, SPECTRIN ALPHA CHAIN, SULFATE ION | | Authors: | Vega, M.C, Viguera, A.R, Serrano, L. | | Deposit date: | 2000-11-06 | | Release date: | 2001-11-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Unspecific Hydrophobic Stabilization of Folding Transition States
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1GCQ
 
 | | CRYSTAL STRUCTURE OF VAV AND GRB2 SH3 DOMAINS | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, GROWTH FACTOR RECEPTOR-BOUND PROTEIN 2, VAV PROTO-ONCOGENE | | Authors: | Nishida, M, Nagata, K, Hachimori, Y, Ogura, K, Inagaki, F. | | Deposit date: | 2000-08-08 | | Release date: | 2001-08-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Novel recognition mode between Vav and Grb2 SH3 domains.
EMBO J., 20, 2001
|
|
1H3H
 
 | | Structural Basis for Specific Recognition of an RxxK-containing SLP-76 peptide by the Gads C-terminal SH3 domain | | Descriptor: | GRB2-RELATED ADAPTOR PROTEIN 2, LYMPHOCYTE CYTOSOLIC PROTEIN 2 | | Authors: | Liu, Q, Berry, D, Nash, P, Pawson, T, McGlade, C.J, Li, S.S. | | Deposit date: | 2002-09-03 | | Release date: | 2003-03-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Specific Binding of the Gads SH3 Domain to an Rxxk Motif-Containing Slp-76 Peptide: A Novel Mode of Peptide Recognition
Mol.Cell, 11, 2003
|
|
1GFD
 
 | |
1GBQ
 
 | | SOLUTION NMR STRUCTURE OF THE GRB2 N-TERMINAL SH3 DOMAIN COMPLEXED WITH A TEN-RESIDUE PEPTIDE DERIVED FROM SOS DIRECT REFINEMENT AGAINST NOES, J-COUPLINGS, AND 1H AND 13C CHEMICAL SHIFTS, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | GRB2, SOS-1 | | Authors: | Wittekind, M, Mapelli, C, Lee, V, Goldfarb, V, Friedrichs, M.S, Meyers, C.A, Mueller, L. | | Deposit date: | 1996-12-23 | | Release date: | 1997-09-04 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Grb2 N-terminal SH3 domain complexed with a ten-residue peptide derived from SOS: direct refinement against NOEs, J-couplings and 1H and 13C chemical shifts.
J.Mol.Biol., 267, 1997
|
|
1GCP
 
 | | CRYSTAL STRUCTURE OF VAV SH3 DOMAIN | | Descriptor: | VAV PROTO-ONCOGENE | | Authors: | Nishida, M, Nagata, K, Hachimori, Y, Ogura, K, Inagaki, F. | | Deposit date: | 2000-08-08 | | Release date: | 2001-08-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Novel recognition mode between Vav and Grb2 SH3 domains.
EMBO J., 20, 2001
|
|
1H8K
 
 | |
1E7O
 
 | | A-SPECTRIN SH3 DOMAIN A11V, V23L, M25V, V44I, V58L MUTATIONS | | Descriptor: | GLYCEROL, SPECTRIN ALPHA CHAIN | | Authors: | Vega, M.C, Serrano, L. | | Deposit date: | 2000-08-31 | | Release date: | 2003-05-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | A Thermodynamic and Kinetic Analysis of the Folding Pathway of an SH3 Domain Entropically Stabilised by a Redesigned Hydrophobic Core
J.Mol.Biol., 328, 2003
|
|
1H92
 
 | | SH3 domain of human Lck tyrosine kinase | | Descriptor: | PROTO-ONCOGENE TYROSINE-PROTEIN KINASE LCK | | Authors: | Schweimer, K, Hoffmann, S, Friedrich, U, Biesinger, B, Roesch, P, Sticht, H. | | Deposit date: | 2001-02-22 | | Release date: | 2001-10-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Investigation of the Binding of a Herpesviral Protein to the SH3 Domain of Tyrosine Kinase Lck
Biochemistry, 41, 2002
|
|
1HSQ
 
 | | SOLUTION STRUCTURE OF THE SH3 DOMAIN OF PHOSPHOLIPASE CGAMMA | | Descriptor: | PHOSPHOLIPASE C-GAMMA (SH3 DOMAIN) | | Authors: | Kohda, D, Hatanaka, H, Odaka, M, Inagaki, F. | | Deposit date: | 1994-06-13 | | Release date: | 1994-08-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the SH3 domain of phospholipase C-gamma.
Cell(Cambridge,Mass.), 72, 1993
|
|
1I07
 
 | | EPS8 SH3 DOMAIN INTERTWINED DIMER | | Descriptor: | EPIDERMAL GROWTH FACTOR RECEPTOR KINASE SUBSTRATE EPS8 | | Authors: | Kishan, K.V.R, Newcomer, M.E. | | Deposit date: | 2001-01-29 | | Release date: | 2001-05-09 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Effect of pH and salt bridges on structural assembly: molecular structures of the monomer and intertwined dimer of the Eps8 SH3 domain.
Protein Sci., 10, 2001
|
|
1I0C
 
 | | EPS8 SH3 CLOSED MONOMER | | Descriptor: | EPIDERMAL GROWTH FACTOR RECEPTOR KINASE SUBSTRATE EPS8 | | Authors: | Kishan, K.V.R, Newcomer, M.E. | | Deposit date: | 2001-01-29 | | Release date: | 2001-05-09 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Effect of pH and salt bridges on structural assembly: molecular structures of the monomer and intertwined dimer of the Eps8 SH3 domain.
Protein Sci., 10, 2001
|
|
1IO6
 
 | |
1K1Z
 
 | | Solution structure of N-terminal SH3 domain mutant(P33G) of murine Vav | | Descriptor: | vav | | Authors: | Ogura, K, Nagata, K, Horiuchi, M, Ebisui, E, Hasuda, T, Yuzawa, S, Nishida, M, Hatanaka, H, Inagaki, F. | | Deposit date: | 2001-09-26 | | Release date: | 2001-10-10 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of N-terminal SH3 domain of Vav and the recognition site for Grb2 C-terminal SH3 domain
J.BIOMOL.NMR, 22, 2002
|
|
1JQQ
 
 | | Crystal structure of Pex13p(301-386) SH3 domain | | Descriptor: | PEROXISOMAL MEMBRANE PROTEIN PAS20 | | Authors: | Douangamath, A, Mayans, O, Barnett, P, Distel, B, Wilmanns, M. | | Deposit date: | 2001-08-08 | | Release date: | 2002-12-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Topography for Independent Binding of alpha-Helical and PPII-Helical Ligands to a Peroxisomal SH3 Domain
Mol.Cell, 10, 2002
|
|
1KFZ
 
 | | Solution Structure of C-terminal Sem-5 SH3 Domain (Ensemble of 16 Structures) | | Descriptor: | SEX MUSCLE ABNORMAL PROTEIN 5 | | Authors: | Ferreon, J.C, Volk, D.E, Luxon, B.A, Gorenstein, D, Hilser, V.J. | | Deposit date: | 2001-11-24 | | Release date: | 2003-05-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure, Dynamics and Thermodynamics of the Native State Ensemble
of Sem-5 C-Terminal SH3 Domain
Biochemistry, 42, 2003
|
|
1K76
 
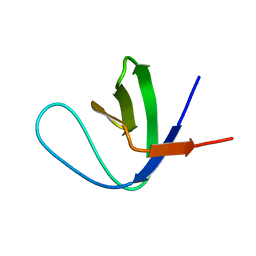 | | Solution Structure of the C-terminal Sem-5 SH3 Domain (Minimized Average Structure) | | Descriptor: | SEX MUSCLE ABNORMAL PROTEIN 5 | | Authors: | Ferreon, J, Volk, D, Luxon, B, Gorenstein, D, Hilser, V. | | Deposit date: | 2001-10-18 | | Release date: | 2003-05-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure, Dynamics and Thermodynamics of the Native State Ensemble
of Sem-5 C-Terminal SH3 Domain
Biochemistry, 42, 2003
|
|
1KIK
 
 | | SH3 Domain of Lymphocyte Specific Kinase (LCK) | | Descriptor: | PROTO-ONCOGENE TYROSINE-PROTEIN KINASE LCK | | Authors: | Briese, L, Willbold, D. | | Deposit date: | 2001-12-03 | | Release date: | 2001-12-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure determination of human Lck unique and SH3 domains by nuclear magnetic resonance spectroscopy.
Bmc Struct.Biol., 3, 2003
|
|
1JEG
 
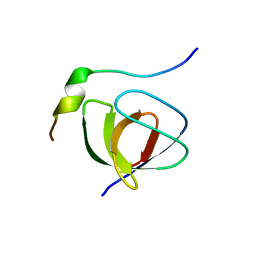 | | Solution structure of the SH3 domain from C-terminal Src Kinase complexed with a peptide from the tyrosine phosphatase PEP | | Descriptor: | HEMATOPOIETIC CELL PROTEIN-TYROSINE PHOSPHATASE 70Z-PEP, TYROSINE-PROTEIN KINASE CSK | | Authors: | Ghose, R, Shekhtman, A, Goger, M.J, Ji, H, Cowburn, D. | | Deposit date: | 2001-06-17 | | Release date: | 2001-10-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A novel, specific interaction involving the Csk SH3 domain and its natural ligand.
Nat.Struct.Biol., 8, 2001
|
|
1JO8
 
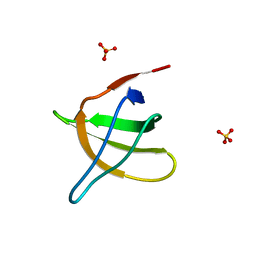 | | Structural analysis of the yeast actin binding protein Abp1 SH3 domain | | Descriptor: | ACTIN BINDING PROTEIN, SULFATE ION | | Authors: | Fazi, B, Cope, M.J, Douangamath, A, Ferracuti, S, Schirwitz, K, Zucconi, A, Drubin, D.G, Wilmanns, M, Cesareni, G, Castagnoli, L. | | Deposit date: | 2001-07-27 | | Release date: | 2002-03-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Unusual binding properties of the SH3 domain of the yeast actin-binding protein Abp1: structural and functional analysis.
J.Biol.Chem., 277, 2002
|
|
1M30
 
 | | Solution structure of N-terminal SH3 domain from oncogene protein c-Crk | | Descriptor: | Proto-oncogene C-crk | | Authors: | Schumann, F.H, Varadan, R, Tayakuniyil, P.P, Hall, J.B, Camarero, J.A, Fushman, D. | | Deposit date: | 2002-06-26 | | Release date: | 2003-08-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Changing protein backbone topology: Structural and dynamic consequences of the backbone cyclization in SH3 domain
To be Published
|
|
1M3C
 
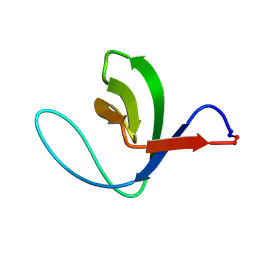 | | Solution structure of a circular form of the N-terminal SH3 domain (E132C, E133G, R191G mutant) from oncogene protein c-Crk | | Descriptor: | Proto-oncogene C-crk | | Authors: | Schumann, F.H, Varadan, R, Tayakuniyil, P.P, Hall, J.B, Camarero, J.A, Fushman, D. | | Deposit date: | 2002-06-27 | | Release date: | 2003-08-05 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | Changing protein backbone topology: Structural and dynamic consequences of the backbone cyclization in SH3 domain
To be Published
|
|
1M3B
 
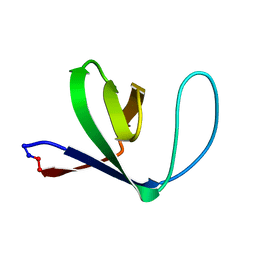 | | Solution structure of a circular form of the N-terminal SH3 domain (A134C, E135G, R191G mutant) from oncogene protein c-Crk. | | Descriptor: | Proto-oncogene C-crk | | Authors: | Schumann, F.H, Varadan, R, Tayakuniyil, P.P, Hall, J.B, Camarero, J.A, Fushman, D. | | Deposit date: | 2002-06-27 | | Release date: | 2003-08-05 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | Changing protein backbone topology: Structural and dynamic consequences of the backbone cyclization in SH3 domain
To be Published
|
|
