4D9S
 
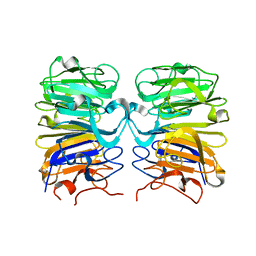 | | Crystal structure of Arabidopsis thaliana UVR8 (UV Resistance locus 8) | | Descriptor: | UVB-resistance protein UVR8 | | Authors: | Arvai, A.S, Christie, J.M, Pratt, A.J, Hitomi, K, Getzoff, E.D. | | Deposit date: | 2012-01-11 | | Release date: | 2012-04-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Plant UVR8 Photoreceptor Senses UV-B by Tryptophan-Mediated Disruption of Cross-Dimer Salt Bridges.
Science, 335, 2012
|
|
2J4D
 
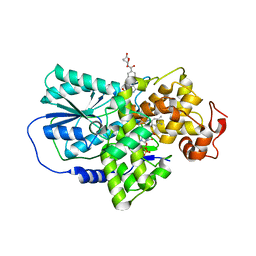 | | Cryptochrome 3 from Arabidopsis thaliana | | Descriptor: | 5,10-METHENYL-6,7,8-TRIHYDROFOLIC ACID, CRYPTOCHROME DASH, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Klar, T, Pokorny, R, Batschauer, A, Essen, L.-O. | | Deposit date: | 2006-08-28 | | Release date: | 2007-06-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Cryptochrome 3 from Arabidopsis Thaliana: Structural and Functional Analysis of its Complex with a Folate Light Antenna
J.Mol.Biol., 366, 2007
|
|
2J09
 
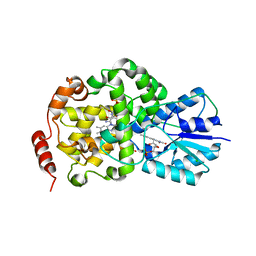 | | Thermus DNA photolyase with FMN antenna chromophore | | Descriptor: | CHLORIDE ION, DEOXYRIBODIPYRIMIDINE PHOTO-LYASE, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Klar, T, Kaiser, G, Hennecke, U, Carell, T, Batschauer, A, Essen, L.-O. | | Deposit date: | 2006-08-01 | | Release date: | 2007-05-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Natural and Non-Natural Antenna Chromophores in the DNA Photolyase from Thermus Thermophilus
Chembiochem, 7, 2006
|
|
2J07
 
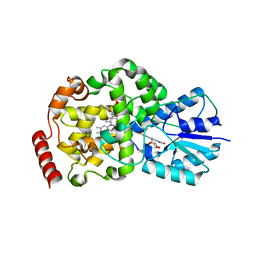 | | Thermus DNA photolyase with 8-HDF antenna chromophore | | Descriptor: | 8-HYDROXY-10-(D-RIBO-2,3,4,5-TETRAHYDROXYPENTYL)-5-DEAZAISOALLOXAZINE, CHLORIDE ION, DEOXYRIBODIPYRIMIDINE PHOTO-LYASE, ... | | Authors: | Klar, T, Kaiser, G, Hennecke, U, Carell, T, Batschauer, A, Essen, L.-O. | | Deposit date: | 2006-08-01 | | Release date: | 2007-05-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Natural and Non-Natural Antenna Chromophores in the DNA Photolyase from Thermus Thermophilus
Chembiochem, 7, 2006
|
|
2J08
 
 | | Thermus DNA photolyase with 8-Iod-riboflavin antenna chromophore | | Descriptor: | 1-DEOXY-1-(8-IODO-7-METHYL-2,4-DIOXO-3,4-DIHYDROBENZO[G]PTERIDIN-10(2H)-YL)-D-RIBITOL, CHLORIDE ION, DEOXYRIBODIPYRIMIDINE PHOTO-LYASE, ... | | Authors: | Klar, T, Kaiser, G, Hennecke, U, Carell, T, Batschauer, A, Essen, L.-O. | | Deposit date: | 2006-08-01 | | Release date: | 2007-05-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Natural and Non-Natural Antenna Chromophores in the DNA Photolyase from Thermus Thermophilus
Chembiochem, 7, 2006
|
|
2K6A
 
 | | Solution structure of EAS D15 truncation mutant | | Descriptor: | Hydrophobin | | Authors: | Kwan, A.H. | | Deposit date: | 2008-07-07 | | Release date: | 2008-08-19 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | The Cys3-Cys4 loop of the hydrophobin EAS is not required for rodlet formation and surface activity.
J.Mol.Biol., 382, 2008
|
|
2LFN
 
 | | Identification of the key regions that drive functional amyloid formation by the fungal hydrophobin EAS | | Descriptor: | Hydrophobin | | Authors: | Macindoe, I, Kwan, A.H, Morris, V.K, Mackay, J.P, Sunde, M. | | Deposit date: | 2011-07-06 | | Release date: | 2012-01-25 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Self-assembly of functional, amphipathic amyloid monolayers by the fungal hydrophobin EAS
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4G9J
 
 | | Protein Ser/Thr phosphatase-1 in complex with cell-permeable peptide | | Descriptor: | MANGANESE (II) ION, Serine/threonine-protein phosphatase PP1-alpha catalytic subunit, synthetic peptide | | Authors: | Sukackaite, R, Chatterjee, J, Beullens, M, Bollen, M, Koehn, M, Hart, D.J. | | Deposit date: | 2012-07-24 | | Release date: | 2012-09-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Development of a Peptide that Selectively Activates Protein Phosphatase-1 in Living Cells.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
6OBP
 
 | | Reconstituted PP1 holoenzyme | | Descriptor: | CHLORIDE ION, MANGANESE (II) ION, PHOSPHATE ION, ... | | Authors: | Choy, M.S, Moon, T.M, Bray, J.A, Archuleta, T.L, Shi, W, Peti, W, Page, R. | | Deposit date: | 2019-03-21 | | Release date: | 2019-09-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | SDS22 selectively recognizes and traps metal-deficient inactive PP1.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6OBN
 
 | | The crystal structure of coexpressed SDS22:PP1 complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, FE (III) ION, ... | | Authors: | Choy, M.S, Moon, T.M, Bray, J.A, Archuleta, T.L, Shi, W, Peti, W, Page, R. | | Deposit date: | 2019-03-21 | | Release date: | 2019-09-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | SDS22 selectively recognizes and traps metal-deficient inactive PP1.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
3HJI
 
 | |
3HJK
 
 | |
6ZEJ
 
 | | Structure of PP1-Phactr1 chimera [PP1(7-304) + linker (SGSGS) + Phactr1(526-580)] | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Mouilleron, S, Treisman, R, Fedoryshchak, R, Lee, R, Butler, A.M, Prechova, M. | | Deposit date: | 2020-06-16 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Molecular basis for substrate specificity of the Phactr1/PP1 phosphatase holoenzyme.
Elife, 9, 2020
|
|
6ZEI
 
 | | Structure of PP1-IRSp53 S455E chimera [PP1(7-304) + linker (G/S)x9 + IRSp53(449-465)] bound to Phactr1 (516-580) | | Descriptor: | GLYCEROL, MANGANESE (II) ION, PHOSPHATE ION, ... | | Authors: | Mouilleron, S, Treisman, R, Fedoryshchak, R, Lee, R, Butler, A.M, Prechova, M. | | Deposit date: | 2020-06-16 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Molecular basis for substrate specificity of the Phactr1/PP1 phosphatase holoenzyme.
Elife, 9, 2020
|
|
6ZEH
 
 | | Structure of PP1-spectrin alpha II chimera [PP1(7-304) + linker (G/S)x9 + spectrin alpha II (1025-1039)] bound to Phactr1 (516-580) | | Descriptor: | MANGANESE (II) ION, PHOSPHATE ION, Phosphatase and actin regulator, ... | | Authors: | Mouilleron, S, Treisman, R, Fedoryshchak, R, Lee, R, Butler, A.M, Prechova, M. | | Deposit date: | 2020-06-16 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Molecular basis for substrate specificity of the Phactr1/PP1 phosphatase holoenzyme.
Elife, 9, 2020
|
|
3HVQ
 
 | | Crystal structure of a complex between Protein Phosphatase 1 alpha (PP1) and the PP1 binding and PDZ domains of Neurabin | | Descriptor: | GLYCEROL, MANGANESE (II) ION, Neurabin-1, ... | | Authors: | Critton, D.A, Ragusa, M.J, Page, R, Peti, W. | | Deposit date: | 2009-06-16 | | Release date: | 2010-03-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Spinophilin directs protein phosphatase 1 specificity by blocking substrate binding sites.
Nat.Struct.Mol.Biol., 17, 2010
|
|
6ZK6
 
 | | Protein Phosphatase 1 (PP1) T320E mutant | | Descriptor: | FE (III) ION, MANGANESE (II) ION, PHOSPHATE ION, ... | | Authors: | Salvi, F, Barabas, O, Koehn, M. | | Deposit date: | 2020-06-29 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Towards Dissecting the Mechanism of Protein Phosphatase-1 Inhibition by Its C-Terminal Phosphorylation.
Chembiochem, 22, 2021
|
|
6ZEG
 
 | | Structure of PP1-IRSp53 chimera [PP1(7-304) + linker (G/S)x9 + IRSp53(449-465)] bound to Phactr1 (516-580) | | Descriptor: | 1,2-ETHANEDIOL, 3,6,9,12,15,18-HEXAOXAICOSANE, MANGANESE (II) ION, ... | | Authors: | Mouilleron, S, Treisman, R, Fedoryshchak, R, Lee, R, Butler, A.M, Prechova, M. | | Deposit date: | 2020-06-16 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Molecular basis for substrate specificity of the Phactr1/PP1 phosphatase holoenzyme.
Elife, 9, 2020
|
|
2NZ2
 
 | | Crystal structure of human argininosuccinate synthase in complex with aspartate and citrulline | | Descriptor: | ASPARTIC ACID, Argininosuccinate synthase, CITRULLINE, ... | | Authors: | Karlberg, T, Uppenberg, J, Arrowsmith, C, Berglund, H, Busam, R.D, Collins, R, Edwards, A, Ericsson, U.B, Flodin, S, Flores, A, Graslund, S, Hallberg, B.M, Hammarstrom, M, Hogbom, M, Johansson, I, Kotenyova, T, Magnusdottir, A, Moche, M, Nilsson, M.E, Nordlund, P, Nyman, T, Ogg, D, Persson, C, Sagemark, J, Stenmark, P, Sundstrom, M, Thorsell, A.G, Van Den Berg, S, Wallden, K, Weigelt, J, Holmberg-Schiavone, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-11-22 | | Release date: | 2006-12-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of human argininosuccinate synthetase.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
2O8A
 
 | | rat PP1cgamma complexed with mouse inhibitor-2 | | Descriptor: | Protein phosphatase inhibitor 2, Serine/threonine-protein phosphatase PP1-gamma catalytic subunit | | Authors: | Hurley, T.D. | | Deposit date: | 2006-12-12 | | Release date: | 2007-07-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structural basis for regulation of protein phosphatase 1 by inhibitor-2.
J.Biol.Chem., 282, 2007
|
|
2O8G
 
 | | Rat pp1c gamma complexed with mouse inhibitor-2 | | Descriptor: | MANGANESE (II) ION, Protein phosphatase inhibitor 2, Serine/threonine-protein phosphatase PP1-gamma catalytic subunit | | Authors: | Hurley, T.D. | | Deposit date: | 2006-12-12 | | Release date: | 2007-07-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for regulation of protein phosphatase 1 by inhibitor-2.
J.Biol.Chem., 282, 2007
|
|
3IS2
 
 | | 2.3 Angstrom Crystal Structure of a Cys71 Sulfenic Acid form of Vivid | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Vivid PAS protein VVD | | Authors: | Zoltowski, B.D, Lamb, J.S, Pabit, S.A, Li, L, Pollack, L, Crane, B.R. | | Deposit date: | 2009-08-25 | | Release date: | 2009-11-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Illuminating solution responses of a LOV domain protein with photocoupled small-angle X-ray scattering.
J.Mol.Biol., 393, 2009
|
|
7AOA
 
 | | Structure of the extended MTA1/HDAC1/MBD2/RBBP4 NURD deacetylase complex | | Descriptor: | Histone deacetylase 1, Histone-binding protein RBBP4, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Millard, C.J, Fairall, L, Ragan, T.J, Savva, C.G, Schwabe, J.W.R. | | Deposit date: | 2020-10-14 | | Release date: | 2020-11-11 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (19.4 Å) | | Cite: | The topology of chromatin-binding domains in the NuRD deacetylase complex.
Nucleic Acids Res., 48, 2020
|
|
7AO9
 
 | | Structure of the core MTA1/HDAC1/MBD2 NURD deacetylase complex | | Descriptor: | Histone deacetylase 1, INOSITOL HEXAKISPHOSPHATE, Metastasis-associated protein MTA1, ... | | Authors: | Millard, C.J, Fairall, L, Ragan, T.J, Savva, C.G, Schwabe, J.W.R. | | Deposit date: | 2020-10-14 | | Release date: | 2020-11-11 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | The topology of chromatin-binding domains in the NuRD deacetylase complex.
Nucleic Acids Res., 48, 2020
|
|
7AO8
 
 | | Structure of the MTA1/HDAC1/MBD2 NURD deacetylase complex | | Descriptor: | Histone deacetylase 1, INOSITOL HEXAKISPHOSPHATE, Metastasis-associated protein MTA1, ... | | Authors: | Millard, C.J, Fairall, L, Ragan, T.J, Savva, C.G, Schwabe, J.W.R. | | Deposit date: | 2020-10-14 | | Release date: | 2020-11-11 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | The topology of chromatin-binding domains in the NuRD deacetylase complex.
Nucleic Acids Res., 48, 2020
|
|
