1KQ9
 
 | | Human methionine aminopeptidase type II in complex with L-methionine | | Descriptor: | METHIONINE, Methionine aminopeptidase 2, TERTIARY-BUTYL ALCOHOL, ... | | Authors: | Nonato, M.C, Widom, J, Clardy, J. | | Deposit date: | 2002-01-04 | | Release date: | 2003-12-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Human methionine aminopeptidase type 2 in complex with L- and D-methionine
Bioorg.Med.Chem.Lett., 16, 2006
|
|
1KQA
 
 | |
1KQB
 
 | | Structure of Nitroreductase from E. cloacae complex with inhibitor benzoate | | Descriptor: | BENZOIC ACID, FLAVIN MONONUCLEOTIDE, OXYGEN-INSENSITIVE NAD(P)H NITROREDUCTASE | | Authors: | Haynes, C.A, Koder, R.L, Miller, A.F, Rodgers, D.W. | | Deposit date: | 2002-01-04 | | Release date: | 2002-02-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of nitroreductase in three states: effects of inhibitor binding and reduction.
J.Biol.Chem., 277, 2002
|
|
1KQC
 
 | | Structure of Nitroreductase from E. cloacae Complex with Inhibitor Acetate | | Descriptor: | ACETATE ION, FLAVIN MONONUCLEOTIDE, OXYGEN-INSENSITIVE NAD(P)H NITROREDUCTASE | | Authors: | Haynes, C.A, Koder, R.L, Miller, A.F, Rodgers, D.W. | | Deposit date: | 2002-01-04 | | Release date: | 2002-02-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of nitroreductase in three states: effects of inhibitor binding and reduction.
J.Biol.Chem., 277, 2002
|
|
1KQD
 
 | | Structure of Nitroreductase from E. cloacae Bound with 2e-Reduced Flavin Mononucleotide (FMN) | | Descriptor: | FLAVIN MONONUCLEOTIDE, OXYGEN-INSENSITIVE NAD(P)H NITROREDUCTASE | | Authors: | Haynes, C.A, Koder, R.L, Miller, A.F, Rodgers, D.W. | | Deposit date: | 2002-01-04 | | Release date: | 2002-02-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of nitroreductase in three states: effects of inhibitor binding and reduction.
J.Biol.Chem., 277, 2002
|
|
1KQE
 
 | | Solution structure of a linked shortened gramicidin A in benzene/acetone 10:1 | | Descriptor: | MINI-GRAMICIDIN A | | Authors: | Arndt, H.D, Bockelmann, D, Knoll, A, Lamberth, S, Griesinger, C, Koert, U. | | Deposit date: | 2002-01-05 | | Release date: | 2002-11-27 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Cation Control in Functional Helical Programming: Structures of a D,L-Peptide Ion Channel
Angew.Chem.Int.Ed.Engl., 41, 2002
|
|
1KQF
 
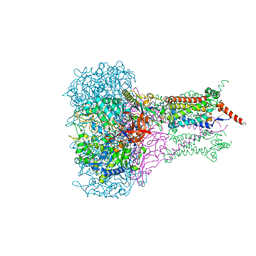 | | FORMATE DEHYDROGENASE N FROM E. COLI | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, CARDIOLIPIN, FORMATE DEHYDROGENASE, ... | | Authors: | Jormakka, M, Tornroth, S, Byrne, B, Iwata, S. | | Deposit date: | 2002-01-05 | | Release date: | 2002-03-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular basis of proton motive force generation: structure of formate dehydrogenase-N.
Science, 295, 2002
|
|
1KQG
 
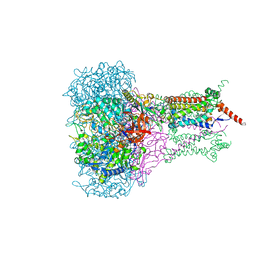 | | FORMATE DEHYDROGENASE N FROM E. COLI | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, 2-HEPTYL-4-HYDROXY QUINOLINE N-OXIDE, CARDIOLIPIN, ... | | Authors: | Jormakka, M, Tornroth, S, Byrne, B, Iwata, S. | | Deposit date: | 2002-01-05 | | Release date: | 2002-03-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular basis of proton motive force generation: structure of formate dehydrogenase-N.
Science, 295, 2002
|
|
1KQH
 
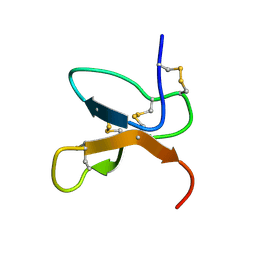 | | NMR Solution Structure of the cis Pro30 Isomer of ACTX-Hi:OB4219 | | Descriptor: | ACTX-Hi:OB4219 | | Authors: | Rosengren, K.J, Wilson, D, Daly, N.L, Alewood, P.F, Craik, D.J. | | Deposit date: | 2002-01-05 | | Release date: | 2002-02-06 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the cis- and trans-Pro30 isomers of a novel 38-residue toxin
from the venom of Hadronyche Infensa sp. that contains a cystine-knot motif within
its four disulfide bonds
Biochemistry, 41, 2002
|
|
1KQI
 
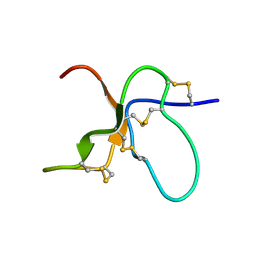 | | NMR Solution Structure of the trans Pro30 Isomer of ACTX-Hi:OB4219 | | Descriptor: | ACTX-Hi:OB4219 | | Authors: | Rosengren, K.J, Wilson, D, Daly, N.L, Alewood, P.F, Craik, D.J. | | Deposit date: | 2002-01-06 | | Release date: | 2002-02-06 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the cis- and trans-Pro30 isomers of a novel 38-residue toxin
from the venom of Hadronyche Infensa sp. that contains a cystine-knot motif within
its four disulfide bonds
Biochemistry, 41, 2002
|
|
1KQJ
 
 | | Crystal Structure of a Mutant of MutY Catalytic Domain | | Descriptor: | A/G-SPECIFIC ADENINE GLYCOSYLASE, GLYCEROL, IRON/SULFUR CLUSTER, ... | | Authors: | Messick, T.E, Chmiel, N.H, Golinelli, M.P, David, S.S, Joshua-Tor, L. | | Deposit date: | 2002-01-06 | | Release date: | 2002-04-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Noncysteinyl coordination to the [4Fe-4S]2+ cluster of the DNA repair adenine glycosylase MutY introduced via site-directed mutagenesis. Structural characterization of an unusual histidinyl-coordinated cluster.
Biochemistry, 41, 2002
|
|
1KQK
 
 | | Solution Structure of the N-terminal Domain of a Potential Copper-translocating P-type ATPase from Bacillus subtilis in the Cu(I)loaded State | | Descriptor: | COPPER (I) ION, POTENTIAL COPPER-TRANSPORTING ATPASE | | Authors: | Banci, L, Bertini, I, Ciofi-Baffoni, S, D'Onofrio, M, Gonnelli, L, Marhuenda-Egea, F.C, Ruiz-Duenas, F.J. | | Deposit date: | 2002-01-07 | | Release date: | 2002-04-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the N-terminal domain of a potential copper-translocating P-type ATPase from Bacillus subtilis in the apo and Cu(I) loaded states.
J.Mol.Biol., 317, 2002
|
|
1KQL
 
 | | Crystal structure of the C-terminal region of striated muscle alpha-tropomyosin at 2.7 angstrom resolution | | Descriptor: | Fusion Protein of and striated muscle alpha-tropomyosin and the GCN4 leucine zipper | | Authors: | Li, Y, Mui, S, Brown, J.H, Strand, J, Reshetnikova, L, Tobacman, L.S, Cohen, C. | | Deposit date: | 2002-01-07 | | Release date: | 2002-05-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The crystal structure of the C-terminal fragment of striated-muscle alpha-tropomyosin reveals a key troponin T recognition site.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1KQM
 
 | | SCALLOP MYOSIN S1-AMPPNP IN THE ACTIN-DETACHED CONFORMATION | | Descriptor: | CALCIUM ION, MAGNESIUM ION, MYOSIN ESSENTIAL LIGHT CHAIN, ... | | Authors: | Himmel, D.M, Gourinath, S, Reshetnikova, L, Shen, Y, Szent-Gyorgyi, G, Cohen, C. | | Deposit date: | 2002-01-07 | | Release date: | 2002-11-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystallographic findings on the
internally uncoupled and near-rigor
states of myosin: Further insights into
the mechanics of the motor
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1KQN
 
 | | Crystal structure of NMN/NaMN adenylyltransferase complexed with NAD | | Descriptor: | NICOTINAMIDE MONONUCLEOTIDE ADENYLYL TRANSFERASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, XENON | | Authors: | Zhou, T, Kurnasov, O, Tomchick, D.R, Binns, D.D, Grishin, N.V, Marquez, V.E, Osterman, A.L, Zhang, H. | | Deposit date: | 2002-01-07 | | Release date: | 2003-01-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of Human Nicotinamide/Nicotonic Acid Mononucleotide Adenylyltransferase. Basis for the dual substrate specificity and activation of the oncolytic agent tiazofurin.
J.Biol.Chem., 277, 2003
|
|
1KQO
 
 | | Crystal structure of NMN/NaMN adenylyltransferase complexed with deamido-NAD | | Descriptor: | NICOTINAMIDE MONONUCLEOTIDE ADENYLYL TRANSFERASE, NICOTINIC ACID ADENINE DINUCLEOTIDE | | Authors: | Zhou, T, Kurnasov, O, Tomchick, D.R, Binns, D.D, Grishin, N.V, Marquez, V.E, Osterman, A.L, Zhang, H. | | Deposit date: | 2002-01-07 | | Release date: | 2003-01-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of Hhuman of Nicotinamide/Nicotinic Acid Mononucleotide Adenylyltransferase.
Basis for the dual substrate specificity and activation of the oncolytic agent tiazofurin.
J.Biol.Chem., 277, 2002
|
|
1KQP
 
 | | NH3-DEPENDENT NAD+ SYNTHETASE FROM BACILLUS SUBTILIS AT 1 A RESOLUTION | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, NH(3)-dependent NAD(+) synthetase, ... | | Authors: | Symersky, J, Devedjiev, Y, Moore, K, Brouillette, C, DeLucas, L. | | Deposit date: | 2002-01-07 | | Release date: | 2002-06-28 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | NH3-dependent NAD+ synthetase from Bacillus subtilis at 1 A resolution.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1KQQ
 
 | | Solution Structure of the Dead ringer ARID-DNA Complex | | Descriptor: | 5'-D(*CP*CP*AP*CP*AP*TP*CP*AP*AP*TP*AP*CP*AP*GP*G)-3', 5'-D(*CP*CP*TP*GP*TP*AP*TP*TP*GP*AP*TP*GP*TP*GP*G)-3', DEAD RINGER PROTEIN | | Authors: | Iwahara, J, Iwahara, M, Daughdrill, G.W, Ford, J, Clubb, R.T. | | Deposit date: | 2002-01-07 | | Release date: | 2002-03-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The structure of the Dead ringer-DNA complex reveals how AT-rich interaction domains (ARIDs) recognize DNA.
EMBO J., 21, 2002
|
|
1KQR
 
 | | Crystal Structure of the Rhesus Rotavirus VP4 Sialic Acid Binding Domain in Complex with 2-O-methyl-alpha-D-N-acetyl neuraminic acid | | Descriptor: | 2-O-methyl-5-N-acetyl-alpha-D-neuraminic acid, GLYCEROL, SULFATE ION, ... | | Authors: | Dormitzer, P.R, Sun, Z.-Y.J, Wagner, G, Harrison, S.C. | | Deposit date: | 2002-01-07 | | Release date: | 2002-03-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Rhesus Rotavirus VP4 Sialic Acid Binding Domain has a Galectin Fold with a
Novel Carbohydrate Binding Site
Embo J., 21, 2002
|
|
1KQS
 
 | | The Haloarcula marismortui 50S Complexed with a Pretranslocational Intermediate in Protein Synthesis | | Descriptor: | 23S RRNA, 5S RRNA, 6-AMINOHEXANOIC ACID, ... | | Authors: | Schmeing, T.M, Seila, A.C, Hansen, J.L, Freeborn, B, Soukup, J.K, Scaringe, S.A, Strobel, S.A, Moore, P.B, Steitz, T.A. | | Deposit date: | 2002-01-07 | | Release date: | 2002-02-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A pre-translocational intermediate in protein synthesis observed in crystals of enzymatically active 50S subunits.
Nat.Struct.Biol., 9, 2002
|
|
1KQU
 
 | | Human phospholipase A2 complexed with a substrate anologue | | Descriptor: | 6-PHENYL-4(R)-(7-PHENYL-HEPTANOYLAMINO)-HEXANOIC ACID, CALCIUM ION, Phospholipase A2, ... | | Authors: | Tyndall, J.D, Martin, J.L. | | Deposit date: | 2002-01-07 | | Release date: | 2003-11-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | D-Tyrosine as a chiral precusor to potent inhibitors of human nonpancreatic secretory phospholipase A2 (IIa) with antiinflammatory activity.
Chembiochem, 4, 2003
|
|
1KQV
 
 | | Family of NMR Solution Structures of Ca Ln Calbindin D9K | | Descriptor: | LANTHANUM (III) ION, VITAMIN D-DEPENDENT CALCIUM-BINDING PROTEIN | | Authors: | Bertini, I, Donaire, A, Jimenez, B, Luchinat, C, Parigi, G, Piccioli, M, Poggi, L. | | Deposit date: | 2002-01-08 | | Release date: | 2002-01-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Paramagnetism-based versus classical constraints: an analysis of the solution structure of Ca Ln calbindin D9k.
J.Biomol.NMR, 21, 2001
|
|
1KQW
 
 | | Crystal structure of holo-CRBP from zebrafish | | Descriptor: | Cellular retinol-binding protein, RETINOL | | Authors: | Calderone, V, Folli, C, Marchesani, A, Berni, R, Zanotti, G. | | Deposit date: | 2002-01-08 | | Release date: | 2002-08-28 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Identification and structural analysis of a zebrafish apo and holo cellular retinol-binding protein.
J.Mol.Biol., 321, 2002
|
|
1KQX
 
 | | Crystal structure of apo-CRBP from zebrafish | | Descriptor: | Cellular retinol-binding protein | | Authors: | Calderone, V, Folli, C, Marchesani, A, Berni, R, Zanotti, G. | | Deposit date: | 2002-01-08 | | Release date: | 2002-08-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Identification and structural analysis of a zebrafish apo and holo cellular retinol-binding protein.
J.Mol.Biol., 321, 2002
|
|
1KQY
 
 | | Hevamine Mutant D125A/E127A/Y183F in Complex with Penta-NAG | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Hevamine A, ... | | Authors: | Rozeboom, H.J, Dijkstra, B.W. | | Deposit date: | 2002-01-08 | | Release date: | 2002-01-23 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Expression and characterization of active site mutants of hevamine, a chitinase from the rubber tree Hevea brasiliensis.
Eur.J.Biochem., 269, 2002
|
|
