5GHL
 
 | | Crystal structure Analysis of the starch-binding domain of glucoamylase from Aspergillus niger | | Descriptor: | GLYCEROL, Glucoamylase, SULFATE ION | | Authors: | Miyake, H, Suyama, Y, Muraki, N, Kusunoki, M, Tanaka, A. | | Deposit date: | 2016-06-20 | | Release date: | 2017-10-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the starch-binding domain of glucoamylase from Aspergillus niger.
Acta Crystallogr.,Sect.F, 73, 2017
|
|
7CZJ
 
 | |
8X8K
 
 | | Crystal structure of STBD1 CBM20 domain in complex with maltotetraose | | Descriptor: | GLYCEROL, Starch-binding domain-containing protein 1, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Zhang, Y.C, Pan, L.F. | | Deposit date: | 2023-11-27 | | Release date: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Decoding the molecular mechanism of selective autophagy of glycogen mediated by autophagy receptor STBD1.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
2Z0B
 
 | | Crystal structure of CBM20 domain of human putative glycerophosphodiester phosphodiesterase 5 (KIAA1434) | | Descriptor: | PHOSPHATE ION, Putative glycerophosphodiester phosphodiesterase 5 | | Authors: | Saijo, S, Nishino, A, Kishishita, S, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-07 | | Release date: | 2008-05-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of CBM20 domain of human putative glycerophosphodiester phosphodiesterase 5 (KIAA1434)
To be Published
|
|
1KUL
 
 | | GLUCOAMYLASE, GRANULAR STARCH-BINDING DOMAIN, NMR, 5 STRUCTURES | | Descriptor: | GLUCOAMYLASE | | Authors: | Sorimachi, K, Jacks, A.J, Le Gal-Coeffet, M.-F, Williamson, G, Archer, D.B, Williamson, M.P. | | Deposit date: | 1996-01-12 | | Release date: | 1996-07-11 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the granular starch binding domain of glucoamylase from Aspergillus niger by nuclear magnetic resonance spectroscopy.
J.Mol.Biol., 259, 1996
|
|
1KUM
 
 | | GLUCOAMYLASE, GRANULAR STARCH-BINDING DOMAIN, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | GLUCOAMYLASE | | Authors: | Sorimachi, K, Jacks, A.J, Le Gal-Coeffet, M.-F, Williamson, G, Archer, D.B, Williamson, M.P. | | Deposit date: | 1996-01-12 | | Release date: | 1996-07-11 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the granular starch binding domain of glucoamylase from Aspergillus niger by nuclear magnetic resonance spectroscopy.
J.Mol.Biol., 259, 1996
|
|
1AC0
 
 | | GLUCOAMYLASE, GRANULAR STARCH-BINDING DOMAIN COMPLEX WITH CYCLODEXTRIN, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | Cycloheptakis-(1-4)-(alpha-D-glucopyranose), GLUCOAMYLASE | | Authors: | Sorimachi, K, Le Gal-Coeffet, M.-F, Williamson, G, Archer, D.B, Williamson, M.P. | | Deposit date: | 1997-02-10 | | Release date: | 1997-07-07 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the granular starch binding domain of Aspergillus niger glucoamylase bound to beta-cyclodextrin.
Structure, 5, 1997
|
|
1ACZ
 
 | | GLUCOAMYLASE, GRANULAR STARCH-BINDING DOMAIN COMPLEX WITH CYCLODEXTRIN, NMR, 5 STRUCTURES | | Descriptor: | Cycloheptakis-(1-4)-(alpha-D-glucopyranose), GLUCOAMYLASE | | Authors: | Sorimachi, K, Le Gal-Coeffet, M.-F, Williamson, G, Archer, D.B, Williamson, M.P. | | Deposit date: | 1997-02-10 | | Release date: | 1997-07-07 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the granular starch binding domain of Aspergillus niger glucoamylase bound to beta-cyclodextrin.
Structure, 5, 1997
|
|
1CQY
 
 | | STARCH BINDING DOMAIN OF BACILLUS CEREUS BETA-AMYLASE | | Descriptor: | BETA-AMYLASE | | Authors: | Yoon, H.J, Hirata, A, Adachi, M, Sekine, A, Utsumi, S, Mikami, B. | | Deposit date: | 1999-08-12 | | Release date: | 1999-08-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of Separated Starch-Binding Domain of Bacillus cereus B-amylase
To be Published
|
|
4RKK
 
 | | Structure of a product bound phosphatase | | Descriptor: | Laforin, PHOSPHATE ION, alpha-D-glucopyranose, ... | | Authors: | Vander Kooi, C.W. | | Deposit date: | 2014-10-13 | | Release date: | 2015-01-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural mechanism of laforin function in glycogen dephosphorylation and lafora disease.
Mol.Cell, 57, 2015
|
|
5BCA
 
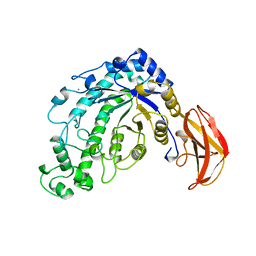 | | BETA-AMYLASE FROM BACILLUS CEREUS VAR. MYCOIDES | | Descriptor: | CALCIUM ION, PROTEIN (1,4-ALPHA-D-GLUCAN MALTOHYDROLASE.) | | Authors: | Oyama, T, Kusunoki, M, Kishimoto, Y, Takasaki, Y, Nitta, Y. | | Deposit date: | 1999-03-12 | | Release date: | 2000-03-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of beta-amylase from Bacillus cereus var. mycoides at 2.2 A resolution.
J.Biochem.(Tokyo), 125, 1999
|
|
6FHV
 
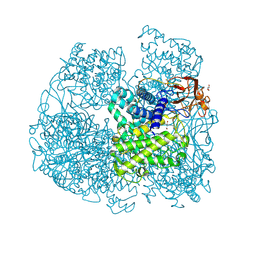 | | Crystal structure of Penicillium oxalicum Glucoamylase | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Roth, C, Moroz, O.V, Ariza, A, Friis, E.P, Davies, G.J, Wilson, K.S. | | Deposit date: | 2018-01-15 | | Release date: | 2018-05-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insight into industrially relevant glucoamylases: flexible positions of starch-binding domains.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
6FHW
 
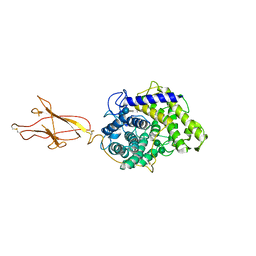 | | Structure of Hormoconis resinae Glucoamylase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Roth, C, Moroz, O.V, Ariza, A, Friis, E.P, Davies, G.J, Wilson, K.S. | | Deposit date: | 2018-01-15 | | Release date: | 2018-05-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural insight into industrially relevant glucoamylases: flexible positions of starch-binding domains.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
8ZRZ
 
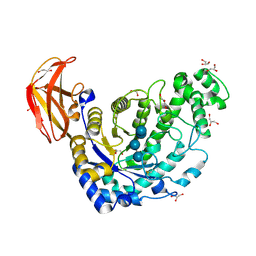 | |
6FRV
 
 | | Structure of the catalytic domain of Aspergillus niger Glucoamylase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glucoamylase, ... | | Authors: | Roth, C, Moroz, O.V, Ariza, A, Friis, E.P, Davies, G.J, Wilson, K.S. | | Deposit date: | 2018-02-16 | | Release date: | 2018-05-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insight into industrially relevant glucoamylases: flexible positions of starch-binding domains.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
1J10
 
 | | beta-amylase from Bacillus cereus var. mycoides in complex with GGX | | Descriptor: | Beta-amylase, CALCIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-xylopyranose, ... | | Authors: | Oyama, T, Miyake, H, Kusunoki, M, Nitta, Y. | | Deposit date: | 2002-11-25 | | Release date: | 2003-06-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of beta-Amylase from Bacillus cereus var. mycoides in Complexes with Substrate Analogs and Affinity-Labeling Reagents
J.BIOCHEM.(TOKYO), 133, 2003
|
|
1J11
 
 | | beta-amylase from Bacillus cereus var. mycoides in complex with alpha-EPG | | Descriptor: | (2R)-oxiran-2-ylmethyl alpha-D-glucopyranoside, Beta-amylase, CALCIUM ION | | Authors: | Oyama, T, Miyake, H, Kusunoki, M, Nitta, Y. | | Deposit date: | 2002-11-25 | | Release date: | 2003-06-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of beta-Amylase from Bacillus cereus var. mycoides in Complexes with Substrate Analogs and Affinity-Labeling Reagents
J.BIOCHEM.(TOKYO), 133, 2003
|
|
1J18
 
 | | Crystal Structure of a Beta-Amylase from Bacillus cereus var. mycoides Cocrystallized with Maltose | | Descriptor: | ACETIC ACID, Beta-amylase, CALCIUM ION, ... | | Authors: | Miyake, H, Kurisu, G, Kusunoki, M, Nishimura, S, Kitamura, S, Nitta, Y. | | Deposit date: | 2002-12-02 | | Release date: | 2003-05-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of a Catalytic Site Mutant of beta-Amylase from Bacillus cereus var. mycoides Cocrystallized with Maltopentaose
BIOCHEMISTRY, 42, 2003
|
|
1ITC
 
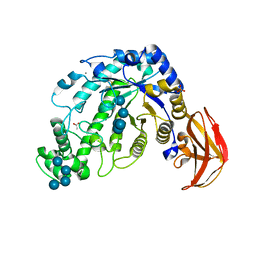 | | Beta-Amylase from Bacillus cereus var. mycoides Complexed with Maltopentaose | | Descriptor: | ACETIC ACID, Beta-Amylase, CALCIUM ION, ... | | Authors: | Miyake, H, Kurisu, G, Kusunoki, M, Nishimura, S, Kitamura, S, Nitta, Y. | | Deposit date: | 2002-01-17 | | Release date: | 2003-05-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of a Catalytic Site Mutant of beta-Amylase from Bacillus cereus var. mycoides Cocrystallized with Maltopentaose
BIOCHEMISTRY, 42, 2003
|
|
1J12
 
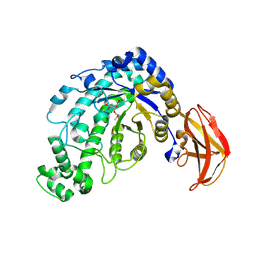 | | Beta-Amylase from Bacillus cereus var. mycoides in Complex with alpha-EBG | | Descriptor: | 2-[(2S)-oxiran-2-yl]ethyl alpha-D-glucopyranoside, Beta-amylase, CALCIUM ION | | Authors: | Oyama, T, Miyake, H, Kusunoki, M, Nitta, Y. | | Deposit date: | 2002-11-25 | | Release date: | 2003-06-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of beta-Amylase from Bacillus cereus var. mycoides in Complexes with Substrate Analogs and Affinity-Labeling Reagents
J.BIOCHEM.(TOKYO), 133, 2003
|
|
1J0Z
 
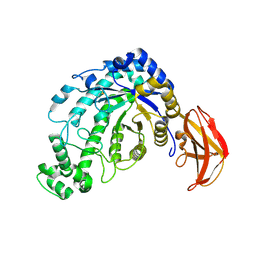 | | Beta-amylase from Bacillus cereus var. mycoides in complex with maltose | | Descriptor: | Beta-amylase, CALCIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Oyama, T, Miyake, H, Kusunoki, M, Nitta, Y. | | Deposit date: | 2002-11-25 | | Release date: | 2003-06-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structures of beta-Amylase from Bacillus cereus var. mycoides in Complexes with Substrate Analogs and Affinity-Labeling Reagents
J.BIOCHEM.(TOKYO), 133, 2003
|
|
1J0Y
 
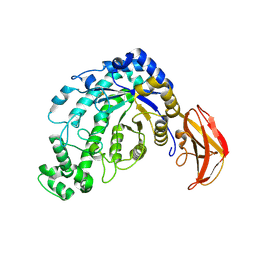 | | Beta-amylase from Bacillus cereus var. mycoides in complex with glucose | | Descriptor: | Beta-amylase, CALCIUM ION, beta-D-glucopyranose | | Authors: | Oyama, T, Miyake, H, Kusunoki, M, Nitta, Y. | | Deposit date: | 2002-11-25 | | Release date: | 2003-06-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of beta-Amylase from Bacillus cereus var. mycoides in Complexes with Substrate Analogs and Affinity-Labeling Reagents
J.BIOCHEM.(TOKYO), 133, 2003
|
|
1B90
 
 | | BACILLUS CEREUS BETA-AMYLASE APO FORM | | Descriptor: | ACETATE ION, CALCIUM ION, PROTEIN (BETA-AMYLASE), ... | | Authors: | Mikami, B, Adachi, M, Kage, T, Sarikaya, E, Nanmori, T, Shinke, R, Utsumi, S. | | Deposit date: | 1999-03-06 | | Release date: | 1999-03-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of raw starch-digesting Bacillus cereus beta-amylase complexed with maltose.
Biochemistry, 38, 1999
|
|
1B9Z
 
 | | BACILLUS CEREUS BETA-AMYLASE COMPLEXED WITH MALTOSE | | Descriptor: | ACETATE ION, CALCIUM ION, PROTEIN (BETA-AMYLASE), ... | | Authors: | Mikami, B, Adachi, M, Kage, T, Sarikaya, E, Nanmori, T, Shinke, R, Utsumi, S. | | Deposit date: | 1999-03-06 | | Release date: | 1999-03-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of raw starch-digesting Bacillus cereus beta-amylase complexed with maltose.
Biochemistry, 38, 1999
|
|
1VEO
 
 | | Crystal Structure Analysis of Y164F/maltose of Bacillus cereus Beta-Amylase at pH 4.6 | | Descriptor: | Beta-amylase, CALCIUM ION, alpha-D-glucopyranose, ... | | Authors: | Hirata, A, Adachi, M, Utsumi, S, Mikami, B. | | Deposit date: | 2004-04-03 | | Release date: | 2005-05-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Engineering of the pH optimum of Bacillus cereus beta-amylase: conversion of the pH optimum from a bacterial type to a higher-plant type
Biochemistry, 43, 2004
|
|
