8HTA
 
 | |
4WBK
 
 | | The 1.37 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with stearic acid | | Descriptor: | Fatty acid-binding protein, heart, STEARIC ACID | | Authors: | Sugiyama, S, Matsuoka, S, Mizohata, E, Matsuoka, D, Murakami, S, Inoue, T, Murata, M. | | Deposit date: | 2014-09-03 | | Release date: | 2015-01-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Molecular Dynamics Simulations of Heart-type Fatty Acid Binding Protein in Apo and Holo Forms, and Hydration Structure Analyses in the Binding Cavity
J.Phys.Chem.B, 119, 2015
|
|
3BY0
 
 | | Crystal structure of Siderocalin (NGAL, Lipocalin 2) W79A-R81A complexed with Ferric Enterobactin | | Descriptor: | 2,3-DIHYDROXY-BENZOIC ACID, 2-(2,3-DIHYDROXY-BENZOYLAMINO)-3-HYDROXY-PROPIONIC ACID, FE (III) ION, ... | | Authors: | Clifton, M.C, Pizzaro, J.C, Strong, R.K. | | Deposit date: | 2008-01-15 | | Release date: | 2008-09-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.572 Å) | | Cite: | The siderocalin/enterobactin interaction: a link between mammalian immunity and bacterial iron transport.
J.Am.Chem.Soc., 130, 2008
|
|
8S1K
 
 | | Crystal Structure of human FABP4 in complex with 2-[1-(methoxymethyl)cyclopentyl]-6-pentyl-4-phenyl-3-(1H-tetrazol-5-yl)-5,6,7,8-tetrahydroquinoline | | Descriptor: | (6~{S})-2-[1-(methoxymethyl)cyclopentyl]-6-pentyl-4-phenyl-3-(1~{H}-1,2,3,4-tetrazol-5-yl)-5,6,7,8-tetrahydroquinoline, DIMETHYL SULFOXIDE, Fatty acid-binding protein, ... | | Authors: | Ehler, A, Benz, J, Obst-Sander, U, Rudolph, M.G. | | Deposit date: | 2024-02-15 | | Release date: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Crystal Structure of a human FABP4 complex
To be published
|
|
7RY5
 
 | | Cellular Retinoic Acid Binding Protein II with Bound Inhibitor 4-[6-({4-[(fluorosulfonyl)oxy]phenyl}ethynyl)-4,4-dimethyl-3,4-dihydroquinolin-1(2H)-yl]-4-oxobutanoic acid | | Descriptor: | 4-[6-({4-[(fluorosulfonyl)oxy]phenyl}ethynyl)-4,4-dimethyl-3,4-dihydroquinolin-1(2H)-yl]-4-oxobutanoic acid, Cellular retinoic acid-binding protein 2 | | Authors: | Kimmel, B.R, Mrksich, M. | | Deposit date: | 2021-08-24 | | Release date: | 2021-12-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Development of an Enzyme-Inhibitor Reaction Using Cellular Retinoic Acid Binding Protein II for One-Pot Megamolecule Assembly.
Chemistry, 27, 2021
|
|
5T43
 
 | |
6VID
 
 | |
1ZRY
 
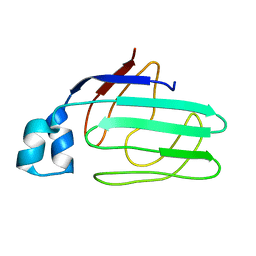 | | NMR structural analysis of apo chicken liver bile acid binding protein | | Descriptor: | Fatty acid-binding protein, liver | | Authors: | Ragona, L, Catalano, M, Luppi, M, Cicero, D, Eliseo, T, Foote, J, Fogolari, F, Zetta, L, Molinari, H. | | Deposit date: | 2005-05-23 | | Release date: | 2006-01-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR Dynamic Studies Suggest that Allosteric Activation Regulates Ligand Binding in Chicken Liver Bile Acid-binding Protein
J.Biol.Chem., 281, 2006
|
|
6VIS
 
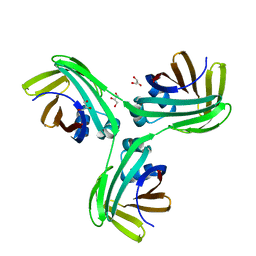 | |
4DQ3
 
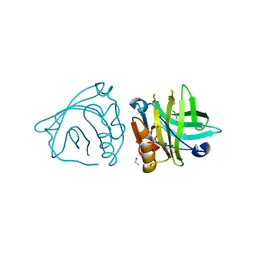 | | Bovine beta-lactoglobulin complex with oleic acid | | Descriptor: | Beta-lactoglobulin, ETHANOL, GLYCEROL, ... | | Authors: | Loch, J.I, Lewinski, K. | | Deposit date: | 2012-02-15 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Binding of 18-carbon unsaturated fatty acids to bovine beta-lactoglobulin--structural and thermodynamic studies.
Int.J.Biol.Macromol., 57, 2013
|
|
1QY0
 
 | | Thermodynamics of Binding of 2-methoxy-3-isopropylpyrazine and 2-methoxy-3-isobutylpyrazine to the Major Urinary Protein | | Descriptor: | CADMIUM ION, GLYCEROL, Major urinary protein, ... | | Authors: | Bingham, R.J, Findlay, J.B.C, Hsieh, S.-Y, Kalverda, A.P, Kjellberg, A, Perazzolo, C, Phillips, S.E.V, Seshadri, K, Trinh, C.H, Turnbull, W.B, Bodenhausen, G, Homans, S.W. | | Deposit date: | 2003-09-09 | | Release date: | 2004-02-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Thermodynamics of Binding of 2-Methoxy-3-isopropylpyrazine and 2-Methoxy-3-isobutylpyrazine to the Major Urinary Protein.
J.Am.Chem.Soc., 126, 2004
|
|
2A0A
 
 | |
4DQ4
 
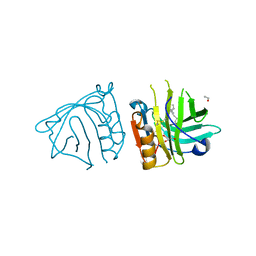 | | Bovine beta-lactoglobulin complex with linoleic acid | | Descriptor: | Beta-lactoglobulin, ETHANOL, GLYCEROL, ... | | Authors: | Loch, J.I, Lewinski, K. | | Deposit date: | 2012-02-15 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Binding of 18-carbon unsaturated fatty acids to bovine beta-lactoglobulin--structural and thermodynamic studies.
Int.J.Biol.Macromol., 57, 2013
|
|
4WFU
 
 | |
4WFV
 
 | |
8IVL
 
 | | FABP7 complexed with Cholesterol | | Descriptor: | CHOLESTEROL, Fatty acid-binding protein, brain | | Authors: | Wei, P.C, Zhao, K, Yin, L. | | Deposit date: | 2023-03-28 | | Release date: | 2024-02-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Fatty acid-binding proteins 3, 7, and 8 bind cholesterol and facilitate its egress from lysosomes.
J.Cell Biol., 223, 2024
|
|
8IVF
 
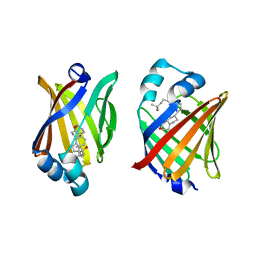 | | FABP7 complexed with 25-HC | | Descriptor: | 25-HYDROXYCHOLESTEROL, Fatty acid-binding protein, brain | | Authors: | Wei, P.C, Zhao, K, Yin, L. | | Deposit date: | 2023-03-27 | | Release date: | 2024-02-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Fatty acid-binding proteins 3, 7, and 8 bind cholesterol and facilitate its egress from lysosomes.
J.Cell Biol., 223, 2024
|
|
2K62
 
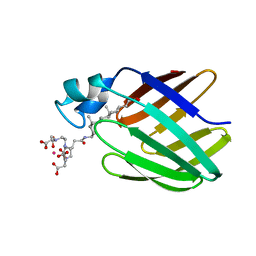 | | NMR solution structure of the supramolecular adduct between a liver cytosolic bile acid binding protein and a bile acid-based Gd(III)-chelate | | Descriptor: | (3alpha,5alpha,8alpha)-3-[(N,N-bis{2-[bis(carboxymethyl)amino]ethyl}-L-gamma-glutamyl)amino]cholan-24-oic acid, Liver fatty acid-binding protein, YTTERBIUM (III) ION | | Authors: | Tomaselli, S, Zanzoni, S, Ragona, L, Gianolio, E, Aime, S, Assfalg, M, Molinari, H. | | Deposit date: | 2008-07-03 | | Release date: | 2008-11-04 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the supramolecular adduct between a liver cytosolic bile acid binding protein and a bile acid-based gadolinium(III)-chelate, a potential hepatospecific magnetic resonance imaging contrast agent.
J.Med.Chem., 51, 2008
|
|
2KT4
 
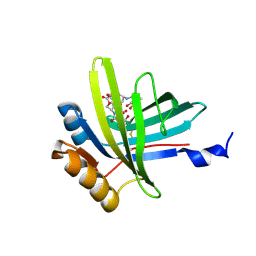 | | Lipocalin Q83 is a Siderocalin | | Descriptor: | Extracellular fatty acid-binding protein, GALLIUM (III) ION, N,N',N''-[(3S,7S,11S)-2,6,10-trioxo-1,5,9-trioxacyclododecane-3,7,11-triyl]tris(2,3-dihydroxybenzamide) | | Authors: | Coudevylle, N, Geist, L, Hartl, M, Kontaxis, G, Bister, K, Konrat, R. | | Deposit date: | 2010-01-18 | | Release date: | 2010-09-08 | | Last modified: | 2016-01-27 | | Method: | SOLUTION NMR | | Cite: | The v-myc-induced Q83 lipocalin is a siderocalin.
J.Biol.Chem., 285, 2010
|
|
2KTD
 
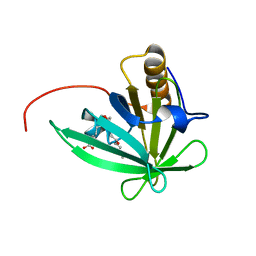 | | Solution structure of mouse lipocalin-type prostaglandin D synthase / substrate analog (U-46619) complex | | Descriptor: | (5Z)-7-{(1R,4S,5S,6R)-6-[(1E,3S)-3-hydroxyoct-1-en-1-yl]-2-oxabicyclo[2.2.1]hept-5-yl}hept-5-enoic acid, Prostaglandin-H2 D-isomerase | | Authors: | Shimamoto, S, Maruo, H, Yoshida, T, Kato, N, Ohkubo, T. | | Deposit date: | 2010-01-27 | | Release date: | 2011-02-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Lipocalin-type Prostaglandin D synthase / Substrate analog complex reveals Open-Closed Conformational Change required for Substrate Recognition
To be Published
|
|
6STS
 
 | | Human myelin protein P2 mutant R30Q | | Descriptor: | Myelin P2 protein, PALMITIC ACID, SULFATE ION | | Authors: | Ruskamo, S, Lehtimaki, M, Kursula, P. | | Deposit date: | 2019-09-11 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Cryo-EM, X-ray diffraction, and atomistic simulations reveal determinants for the formation of a supramolecular myelin-like proteolipid lattice.
J.Biol.Chem., 295, 2020
|
|
5U6G
 
 | |
6T42
 
 | |
8D6H
 
 | | Q108K:K40L:T51C:T53A:R58L:Q38F:Q4F mutant of hCRBPII bound to synthetic fluorophore CM1V after UV irradiation | | Descriptor: | (2E)-3-[7-(diethylamino)-2-oxo-2H-1-benzopyran-3-yl]prop-2-enal, bound form, ACETATE ION, ... | | Authors: | Bingham, C.R, Geiger, J.H, Borhan, B. | | Deposit date: | 2022-06-06 | | Release date: | 2023-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Light controlled reversible Michael addition of cysteine: a new tool for dynamic site-specific labeling of proteins.
Analyst, 148, 2023
|
|
8D6N
 
 | | Q108K:K40L:T53A:R58L:Q38F:Q4F mutant of hCRBPII bound to synthetic fluorophore CM1V | | Descriptor: | (2E)-3-[7-(diethylamino)-2-oxo-2H-1-benzopyran-3-yl]prop-2-enal, bound form, ACETATE ION, ... | | Authors: | Bingham, C.R, Geiger, J.H, Borhan, B. | | Deposit date: | 2022-06-06 | | Release date: | 2023-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Light controlled reversible Michael addition of cysteine: a new tool for dynamic site-specific labeling of proteins.
Analyst, 148, 2023
|
|
