6FEC
 
 | | Human cap-dependent 48S pre-initiation complex | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Schaffitzel, C, Schaffitzel, C. | | Deposit date: | 2017-12-31 | | Release date: | 2018-03-14 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (6.3 Å) | | Cite: | Structure of a human cap-dependent 48S translation pre-initiation complex.
Nucleic Acids Res., 46, 2018
|
|
2I40
 
 | | Cdk2/Cyclin A complexed with a thiophene carboxamide inhibitor | | Descriptor: | 5-[5,6-BIS(METHYLOXY)-1H-BENZIMIDAZOL-1-YL]-3-{[1-(2-CHLOROPHENYL)ETHYL]OXY}-2-THIOPHENECARBOXAMIDE, Cell division protein kinase 2, Cyclin-A2 | | Authors: | Shewchuk, L.M. | | Deposit date: | 2006-08-21 | | Release date: | 2006-10-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | 5-(1H-Benzimidazol-1-yl)-3-alkoxy-2-thiophenecarbonitriles as potent, selective, inhibitors of IKK-epsilon kinase
Bioorg.Med.Chem.Lett., 16, 2006
|
|
8K62
 
 | | Crystal structure of ALKBH1 and 13h complex. | | Descriptor: | 1-[5-[[3-(trifluoromethyloxy)phenyl]methoxy]pyrimidin-2-yl]pyrazole-4-carboxylic acid, MANGANESE (II) ION, Nucleic acid dioxygenase ALKBH1 | | Authors: | Liang, X, Yinping, G, Feng, L, Jiang, Z, Ke, X, Shengyong, Y. | | Deposit date: | 2023-07-24 | | Release date: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.991 Å) | | Cite: | Crystal structure of ALKBH1 and 13h complex
To Be Published
|
|
6MSJ
 
 | | Cryo-EM structures and dynamics of substrate-engaged human 26S proteasome | | Descriptor: | 26S proteasome complex subunit SEM1, 26S proteasome non-ATPase regulatory subunit 1, 26S proteasome non-ATPase regulatory subunit 11, ... | | Authors: | Mao, Y.D. | | Deposit date: | 2018-10-16 | | Release date: | 2018-11-21 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures and dynamics of substrate-engaged human 26S proteasome.
Nature, 565, 2019
|
|
6MSG
 
 | | Cryo-EM structures and dynamics of substrate-engaged human 26S proteasome | | Descriptor: | 26S proteasome complex subunit SEM1, 26S proteasome non-ATPase regulatory subunit 1, 26S proteasome non-ATPase regulatory subunit 11, ... | | Authors: | Mao, Y.D. | | Deposit date: | 2018-10-16 | | Release date: | 2018-11-21 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structures and dynamics of substrate-engaged human 26S proteasome.
Nature, 565, 2019
|
|
6MSB
 
 | | Cryo-EM structures and dynamics of substrate-engaged human 26S proteasome | | Descriptor: | 26S proteasome complex subunit SEM1, 26S proteasome non-ATPase regulatory subunit 1, 26S proteasome non-ATPase regulatory subunit 11, ... | | Authors: | Mao, Y.D. | | Deposit date: | 2018-10-16 | | Release date: | 2018-12-05 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM structures and dynamics of substrate-engaged human 26S proteasome.
Nature, 565, 2019
|
|
5EAM
 
 | | Crystal structure of human WDR5 in complex with compound 9o | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, SULFATE ION, ... | | Authors: | DONG, A, DOMBROVSKI, L, SMIL, D, GETLIK, M, BOLSHAN, Y, WALKER, J.R, SENISTERRA, G, PODA, G, AL-AWAR, R, SCHAPIRA, M, VEDADI, M, Bountra, C, Edwards, A.M, Arrowsmith, C.H, BROWN, P.J, WU, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-10-16 | | Release date: | 2015-11-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-Based Optimization of a Small Molecule Antagonist of the Interaction Between WD Repeat-Containing Protein 5 (WDR5) and Mixed-Lineage Leukemia 1 (MLL1).
J. Med. Chem., 59, 2016
|
|
8K4E
 
 | |
8K3O
 
 | |
4WLQ
 
 | | Crystal structure of mUCH37-hRPN13 CTD complex | | Descriptor: | Proteasomal ubiquitin receptor ADRM1, Ubiquitin carboxyl-terminal hydrolase isozyme L5 | | Authors: | Hemmis, C.W, Hill, C.P, VanderLinden, R, Whitby, F.G. | | Deposit date: | 2014-10-07 | | Release date: | 2015-03-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural Basis for the Activation and Inhibition of the UCH37 Deubiquitylase.
Mol.Cell, 57, 2015
|
|
6MSK
 
 | | Cryo-EM structures and dynamics of substrate-engaged human 26S proteasome | | Descriptor: | 26S proteasome complex subunit SEM1, 26S proteasome non-ATPase regulatory subunit 1, 26S proteasome non-ATPase regulatory subunit 11, ... | | Authors: | Mao, Y.D. | | Deposit date: | 2018-10-16 | | Release date: | 2018-11-21 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures and dynamics of substrate-engaged human 26S proteasome.
Nature, 565, 2019
|
|
6MSH
 
 | | Cryo-EM structures and dynamics of substrate-engaged human 26S proteasome | | Descriptor: | 26S proteasome complex subunit SEM1, 26S proteasome non-ATPase regulatory subunit 1, 26S proteasome non-ATPase regulatory subunit 11, ... | | Authors: | Mao, Y.D. | | Deposit date: | 2018-10-16 | | Release date: | 2018-11-21 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structures and dynamics of substrate-engaged human 26S proteasome.
Nature, 565, 2019
|
|
6MSE
 
 | | Cryo-EM structures and dynamics of substrate-engaged human 26S proteasome | | Descriptor: | 26S proteasome complex subunit SEM1, 26S proteasome non-ATPase regulatory subunit 1, 26S proteasome non-ATPase regulatory subunit 11, ... | | Authors: | Mao, Y.D. | | Deposit date: | 2018-10-16 | | Release date: | 2018-11-21 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures and dynamics of substrate-engaged human 26S proteasome.
Nature, 565, 2019
|
|
2JGZ
 
 | | Crystal structure of phospho-CDK2 in complex with Cyclin B | | Descriptor: | CELL DIVISION PROTEIN KINASE 2, G2/MITOTIC-SPECIFIC CYCLIN-B1 | | Authors: | Brown, N.R, Petri, E, Lowe, E.D, Skamnaki, V, Johnson, L.N. | | Deposit date: | 2007-02-17 | | Release date: | 2007-05-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Cyclin B and cyclin A confer different substrate recognition properties on CDK2.
Cell Cycle, 6, 2007
|
|
6MSD
 
 | | Cryo-EM structures and dynamics of substrate-engaged human 26S proteasome | | Descriptor: | 26S proteasome complex subunit SEM1, 26S proteasome non-ATPase regulatory subunit 1, 26S proteasome non-ATPase regulatory subunit 11, ... | | Authors: | Mao, Y.D. | | Deposit date: | 2018-10-16 | | Release date: | 2018-12-05 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures and dynamics of substrate-engaged human 26S proteasome.
Nature, 565, 2019
|
|
7WJG
 
 | | Streptococcus mutans BusR transcriptional factor | | Descriptor: | CITRIC ACID, Putative transcriptional regulator (GntR family) | | Authors: | Li, X, Liu, N. | | Deposit date: | 2022-01-06 | | Release date: | 2023-01-11 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structural study of transcriptional factor BusR
To Be Published
|
|
5FFO
 
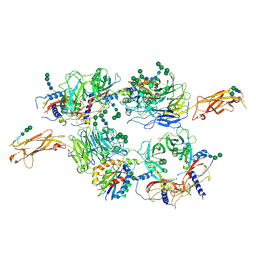 | | Integrin alpha V beta 6 in complex with pro-TGF-beta | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Dong, X, Zhao, B, Springer, T.A. | | Deposit date: | 2015-12-18 | | Release date: | 2017-01-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.49 Å) | | Cite: | Force interacts with macromolecular structure in activation of TGF-beta.
Nature, 542, 2017
|
|
4U50
 
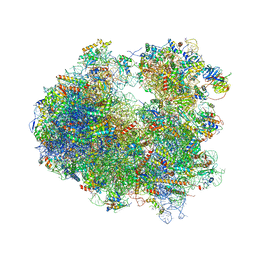 | | Crystal structure of Verrucarin bound to the yeast 80S ribosome | | Descriptor: | (4S,5R,10E,12Z,16R,16aS,17S,18R,19aR,23aR)-4-hydroxy-5,16a,21-trimethyl-4,5,6,7,16,16a,22,23-octahydro-3H,18H,19aH-spiro[16,18-methano[1,6,12]trioxacyclooctadecino[3,4-d]chromene-17,2'-oxirane]-3,9,14-trione, 18S ribosomal RNA, 25S ribosomal RNA, ... | | Authors: | Garreau de Loubresse, N, Prokhorova, I, Yusupova, G, Yusupov, M. | | Deposit date: | 2014-07-24 | | Release date: | 2014-10-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for the inhibition of the eukaryotic ribosome.
Nature, 513, 2014
|
|
7ZKF
 
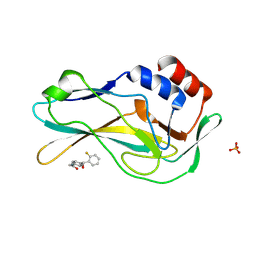 | | Crystal Structure of human Brachyury G177D variant in complex with CF-5-86 | | Descriptor: | PHOSPHATE ION, T-box transcription factor T, UNKNOWN LIGAND | | Authors: | Newman, J.A, Gavard, A, Aitkenhead, H, Imprachim, N, Sherestha, L, Burgess-Brown, N.A, von Delft, F, Bountra, C, Gileadi, O. | | Deposit date: | 2022-04-12 | | Release date: | 2022-06-22 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Crystal Structure of human Brachyury G177D variant in complex with CF-5-86
To Be Published
|
|
7ZL2
 
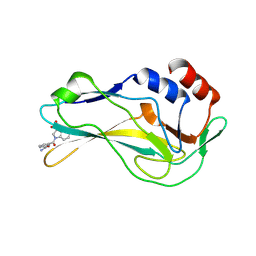 | | Crystal Structure of human Brachyury G177D variant in complex with Molpolrt-039-246-810 | | Descriptor: | T-box transcription factor T, UNKNOWN LIGAND | | Authors: | Newman, J.A, Gavard, A, Aitkenhead, H, Imprachim, N, Sherestha, L, Burgess-Brown, N.A, von Delft, F, Bountra, C, Gileadi, O. | | Deposit date: | 2022-04-13 | | Release date: | 2022-06-22 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of human Brachyury G177D variant in complex with Molpolrt-039-246-810
To Be Published
|
|
7ZK2
 
 | | Crystal Structure of human Brachyury G177D variant in complex with CSC027898502 | | Descriptor: | N-[4-(2-morpholin-4-yl-1,3-thiazol-4-yl)phenyl]ethanamide, PHOSPHATE ION, T-box transcription factor T | | Authors: | Newman, J.A, Gavard, A, Aitkenhead, H, Imprachim, N, Sherestha, L, Burgess-Brown, N.A, von Delft, F, Bountra, C, Gileadi, O. | | Deposit date: | 2022-04-12 | | Release date: | 2022-06-22 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of human Brachyury G177D variant in complex with CSC027898502
To Be Published
|
|
2XGD
 
 | | Crystal structure of a designed homodimeric variant T-A(L)A(L) of the tetracycline repressor | | Descriptor: | CHLORIDE ION, TETRACYCLINE REPRESSOR PROTEIN CLASS B FROM TRANSPOSON TN10, TETRACYCLINE REPRESSOR PROTEIN CLASS D | | Authors: | Stiebritz, M.T, Wengrzik, S, Richter, J.P, Muller, Y.A. | | Deposit date: | 2010-06-03 | | Release date: | 2010-09-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Computational Design of a Chain-Specific Tetracycline Repressor Heterodimer.
J.Mol.Biol., 403, 2010
|
|
2XGC
 
 | | Crystal structure of a designed heterodimeric variant T-A(I)B of the tetracycline repressor | | Descriptor: | TETRACYCLINE REPRESSOR PROTEIN CLASS B FROM TRANSPOSON TN10, TETRACYCLINE REPRESSOR PROTEIN CLASS D | | Authors: | Stiebritz, M.T, Wengrzik, S, Richter, J.P, Muller, Y.A. | | Deposit date: | 2010-06-03 | | Release date: | 2010-09-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Computational Design of a Chain-Specific Tetracycline Repressor Heterodimer.
J.Mol.Biol., 403, 2010
|
|
4U4Y
 
 | | Crystal structure of Pactamycin bound to the yeast 80S ribosome | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Garreau de Loubresse, N, Prokhorova, I, Yusupova, G, Yusupov, M. | | Deposit date: | 2014-07-24 | | Release date: | 2014-10-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for the inhibition of the eukaryotic ribosome.
Nature, 513, 2014
|
|
4U56
 
 | | Crystal structure of Blasticidin S bound to the yeast 80S ribosome | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Garreau de Loubresse, N, Prokhorova, I, Yusupova, G, Yusupov, M. | | Deposit date: | 2014-07-24 | | Release date: | 2014-10-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Structural basis for the inhibition of the eukaryotic ribosome.
Nature, 513, 2014
|
|
