3PBF
 
 | | Surfactant Protein-A neck and carbohydrate recognition domain (NCRD) complexed with glycerol | | Descriptor: | CALCIUM ION, GLYCEROL, Pulmonary surfactant-associated protein A | | Authors: | Shang, F, Rynkiewicz, M.J, McCormack, F.X, Wu, H, Cafarella, T.M, Head, J, Seaton, B.A. | | Deposit date: | 2010-10-20 | | Release date: | 2010-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystallographic complexes of surfactant protein A and carbohydrates reveal ligand-induced conformational change.
J.Biol.Chem., 286, 2011
|
|
1OJX
 
 | | Crystal structure of an Archaeal fructose 1,6-bisphosphate aldolase | | Descriptor: | FRUCTOSE-BISPHOSPHATE ALDOLASE CLASS I | | Authors: | Lorentzen, E, Zwart, P, Stark, A, Hensel, R, Siebers, B, Pohl, E. | | Deposit date: | 2003-07-16 | | Release date: | 2003-09-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of an archaeal class I aldolase and the evolution of (betaalpha)8 barrel proteins.
J. Biol. Chem., 278, 2003
|
|
3DLW
 
 | | Antichymotrypsin | | Descriptor: | Alpha-1-antichymotrypsin | | Authors: | Feil, S.C. | | Deposit date: | 2008-06-29 | | Release date: | 2009-07-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Identification and characterization of a misfolded monomeric serpin formed at physiological temperature
J.Mol.Biol., 403, 2010
|
|
3OFK
 
 | | Crystal structure of N-methyltransferase NodS from Bradyrhizobium japonicum WM9 in complex with S-adenosyl-l-homocysteine (SAH) | | Descriptor: | Nodulation protein S, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Cakici, O, Sikorski, M, Stepkowski, T, Bujacz, G, Jaskolski, M. | | Deposit date: | 2010-08-15 | | Release date: | 2010-10-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structures of NodS N-Methyltransferase from Bradyrhizobium japonicum in Ligand-Free Form and as SAH Complex.
J.Mol.Biol., 404, 2010
|
|
1OKS
 
 | | Crystal structure of the measles virus phosphoprotein XD domain | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, RNA POLYMERASE ALPHA SUBUNIT | | Authors: | Johansson, K, Bourhis, J.-M, Campanacci, V, Cambillau, C, Canard, B, Longhi, S. | | Deposit date: | 2003-07-29 | | Release date: | 2003-09-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the Measles Virus Phosphoprotein Domain Responsible for the Induced Folding of the C-Terminal Domain of the Nucleoprotein
J.Biol.Chem., 278, 2003
|
|
3DWI
 
 | | Crystal structure of Mycobacterium tuberculosis CysM, the cysteine synthase B | | Descriptor: | Cysteine synthase B, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Jurgenson, C.T, Burns, K.E, Begley, T.P, Ealick, S.E. | | Deposit date: | 2008-07-22 | | Release date: | 2008-09-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Crystal structure of a sulfur carrier protein complex found in the cysteine biosynthetic pathway of Mycobacterium tuberculosis.
Biochemistry, 47, 2008
|
|
2VNK
 
 | |
3BBT
 
 | | crystal structure of the ErbB4 kinase in complex with lapatinib | | Descriptor: | N-{3-CHLORO-4-[(3-FLUOROBENZYL)OXY]PHENYL}-6-[5-({[2-(METHYLSULFONYL)ETHYL]AMINO}METHYL)-2-FURYL]-4-QUINAZOLINAMINE, Receptor tyrosine-protein kinase erbB-4 | | Authors: | Qiu, C. | | Deposit date: | 2007-11-11 | | Release date: | 2008-02-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mechanism of Activation and Inhibition of the HER4/ErbB4 Kinase.
Structure, 16, 2008
|
|
1OJM
 
 | |
3BEN
 
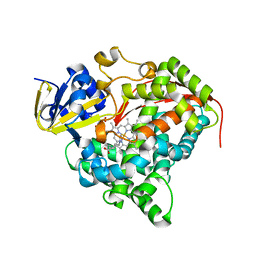 | |
3BFE
 
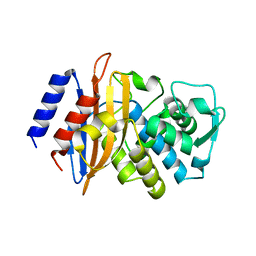 | |
1ONW
 
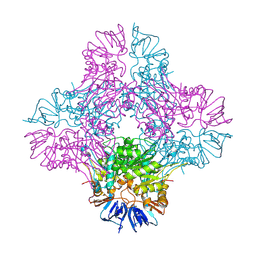 | | Crystal structure of Isoaspartyl Dipeptidase from E. coli | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Isoaspartyl dipeptidase, ... | | Authors: | Thoden, J.B, Marti-Arbona, R, Raushel, F.M, Holden, H.M. | | Deposit date: | 2003-03-02 | | Release date: | 2003-05-06 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | High Resolution X-ray Structure of
Isoaspartyl Dipeptidase from
Escherichia coli
Biochemistry, 42, 2003
|
|
3BFG
 
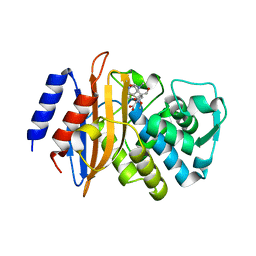 | | class A beta-lactamase SED-G238C complexed with meropenem | | Descriptor: | (4R,5S)-3-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-5-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-4-methyl-4,5-d ihydro-1H-pyrrole-2-carboxylic acid, Class A beta-lactamase Sed1 | | Authors: | Pernot, L, Petrella, S, Sougakoff, W. | | Deposit date: | 2007-11-21 | | Release date: | 2007-12-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | acyl-intermediate structures of the class A beta-lactamase SED-G238C
To be Published
|
|
1OHQ
 
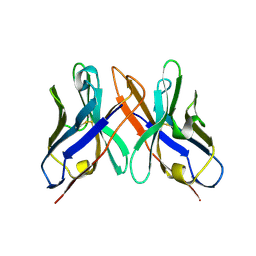 | | Crystal structure of HEL4, a soluble human VH antibody domain resistant to aggregation | | Descriptor: | IMMUNOGLOBULIN | | Authors: | Jespers, L, Schon, O, James, L.C, Veprintsev, D, Winter, G. | | Deposit date: | 2003-05-30 | | Release date: | 2004-03-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Hel4, a Soluble, Refoldable Human V(H) Single Domain with a Germ-Line Scaffold
J.Mol.Biol., 337, 2004
|
|
1F7X
 
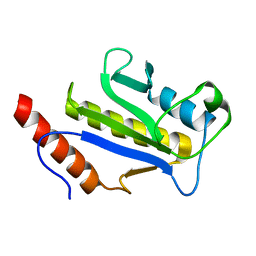 | | SOLUTION STRUCTURE OF C-TERMINAL DOMAIN ZIPA | | Descriptor: | CELL DIVISION PROTEIN ZIPA | | Authors: | Moy, F.J, Glasfeld, E, Mosyak, L, Powers, R. | | Deposit date: | 2000-06-28 | | Release date: | 2001-06-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of ZipA, a crucial component of Escherichia coli cell division.
Biochemistry, 39, 2000
|
|
3O80
 
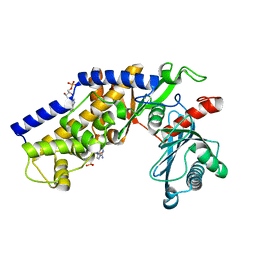 | | Crystal structure of monomeric KlHxk1 in crystal form IX (open state) | | Descriptor: | Hexokinase, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Kuettner, E.B, Kettner, K, Keim, A, Kriegel, T.M, Strater, N. | | Deposit date: | 2010-08-02 | | Release date: | 2010-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Crystal Structure of Hexokinase KlHxk1 of Kluyveromyces lactis: A MOLECULAR BASIS FOR UNDERSTANDING THE CONTROL OF YEAST HEXOKINASE FUNCTIONS VIA COVALENT MODIFICATION AND OLIGOMERIZATION.
J.Biol.Chem., 285, 2010
|
|
1ORE
 
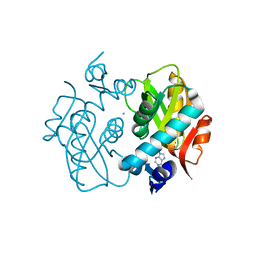 | | Human Adenine Phosphoribosyltransferase | | Descriptor: | ADENOSINE MONOPHOSPHATE, Adenine phosphoribosyltransferase, CHLORIDE ION | | Authors: | Thiemann, O.H, Silva, M, Oliva, G, Silva, C.H.T.P, Iulek, J. | | Deposit date: | 2003-03-12 | | Release date: | 2004-03-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Three-dimensional structure of human adenine phosphoribosyltransferase and its relation to DHA-urolithiasis.
Biochemistry, 43, 2004
|
|
2X3C
 
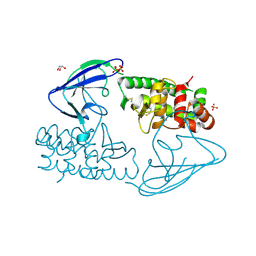 | | AsaP1 inactive mutant E294Q, an extracellular toxic zinc metalloendopeptidase | | Descriptor: | CHLORIDE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Bogdanovic, X, Palm, G.J, Singh, R.K, Hinrichs, W. | | Deposit date: | 2010-01-22 | | Release date: | 2011-02-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural Evidence of Intramolecular Propeptide Inhibition of the Aspzincin Metalloendopeptidase Asap1.
FEBS Lett., 590, 2016
|
|
1OS1
 
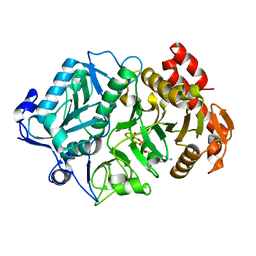 | | Structure of Phosphoenolpyruvate Carboxykinase complexed with ATP,Mg, Ca and pyruvate. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Sudom, A, Walters, R, Pastushok, L, Goldie, D, Prasad, L, Delbaere, L.T, Goldie, H. | | Deposit date: | 2003-03-18 | | Release date: | 2003-09-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mechanisms of activation of phosphoenolpyruvate carboxykinase from Escherichia coli by Ca2+ and of desensitization by trypsin.
J.BACTERIOL., 185, 2003
|
|
2CMA
 
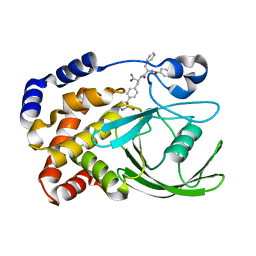 | | Structural Basis for Inhibition of Protein Tyrosine Phosphatase 1B by Isothiazolidinone Heterocyclic Phosphonate Mimetics | | Descriptor: | N-BENZOYL-L-PHENYLALANYL-4-[(5S)-1,1-DIOXIDO-3-OXOISOTHIAZOLIDIN-5-YL]-L-PHENYLALANINAMIDE, TYROSINE-PROTEIN PHOSPHATASE NON-RECEPTOR TYPE 1 | | Authors: | Ala, P.J, Gonneville, L, Hillman, M.C, Becker-Pasha, M, Wei, M, Reid, B.G, Klabe, R, Yue, E.W, Wayland, B, Douty, B, Combs, A.P, Polam, P, Wasserman, Z, Bower, M, Burn, T.C, Hollis, G.F, Wynn, R. | | Deposit date: | 2006-05-04 | | Release date: | 2006-08-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis for Inhibition of Protein-Tyrosine Phosphatase 1B by Isothiazolidinone Heterocyclic Phosphonate Mimetics.
J.Biol.Chem., 281, 2006
|
|
2XAA
 
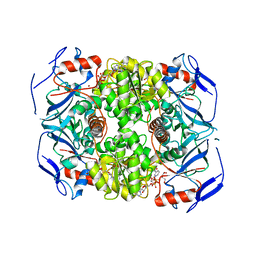 | | Alcohol dehydrogenase ADH-'A' from Rhodococcus ruber DSM 44541 at pH 8.5 in complex with NAD and butane-1,4-diol | | Descriptor: | 1,4-BUTANEDIOL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SECONDARY ALCOHOL DEHYDROGENASE, ... | | Authors: | Kroutil, W, Gruber, K, Grogan, G. | | Deposit date: | 2010-03-30 | | Release date: | 2010-08-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Insights Into Substrate Specificity and Solvent Tolerance in Alcohol Dehydrogenase Adh-'A' from Rhodococcus Ruber Dsm 44541.
Chem.Commun.(Camb.), 46, 2010
|
|
3OFJ
 
 | | Crystal structure of N-methyltransferase NodS from Bradyrhizobium japonicum WM9 | | Descriptor: | Nodulation protein S | | Authors: | Cakici, O, Sikorski, M, Stepkowski, T, Bujacz, G, Jaskolski, M. | | Deposit date: | 2010-08-15 | | Release date: | 2010-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Crystal Structures of NodS N-Methyltransferase from Bradyrhizobium japonicum in Ligand-Free Form and as SAH Complex.
J.Mol.Biol., 404, 2010
|
|
1P4B
 
 | | Three-Dimensional Structure Of a Single Chain Fv Fragment Complexed With The peptide GCN4(7P-14P). | | Descriptor: | Antibody Variable heavy chain, Antibody Variable light chain, GCN4(7P-14P) peptide | | Authors: | Zahnd, C, Spinelli, S, Luginbuhl, B, Jermutus, L, Amstutz, P, Cambillau, C, Pluckthun, A. | | Deposit date: | 2003-04-22 | | Release date: | 2004-05-04 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Directed in Vitro Evolution and Crystallographic Analysis of a Peptide-binding Single Chain Antibody Fragment (scFv) with Low Picomolar Affinity.
J.Biol.Chem., 279, 2004
|
|
2WPE
 
 | | Trypanosoma brucei trypanothione reductase in complex with 3,4- dihydroquinazoline inhibitor (DDD00073359) | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, ... | | Authors: | Alphey, M.S, Patterson, S, Fairlamb, A.H. | | Deposit date: | 2009-08-06 | | Release date: | 2010-10-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Dihydroquinazolines as a Novel Class of Trypanosoma Brucei Trypanothione Reductase Inhibitors: Discovery, Synthesis, and Characterization of Their Binding Mode by Protein Crystallography.
J.Med.Chem., 54, 2011
|
|
2WPC
 
 | | Trypanosoma brucei trypanothione reductase in complex with 3,4- dihydroquinazoline inhibitor (DDD00073357) | | Descriptor: | (4S)-6-CHLORO-3-{2-[4-(FURAN-2-YLCARBONYL)PIPERAZIN-1-YL]ETHYL}-2-METHYL-4-PHENYL-3,4-DIHYDROQUINAZOLINE, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Alphey, M.S, Patterson, S, Fairlamb, A.H. | | Deposit date: | 2009-08-05 | | Release date: | 2010-10-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Dihydroquinazolines as a Novel Class of Trypanosoma Brucei Trypanothione Reductase Inhibitors: Discovery, Synthesis, and Characterization of Their Binding Mode by Protein Crystallography.
J.Med.Chem., 54, 2011
|
|
