5W4K
 
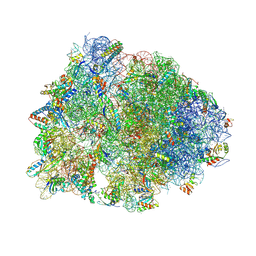 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with Klebsazolicin and bound to mRNA and A-, P- and E-site tRNAs at 2.7A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Metelev, M, Osterman, I.A, Ghilarov, D, Khabibullina, N.F, Yakimov, A, Shabalin, K, Utkina, I, Travin, D.Y, Komarova, E.S, Serebryakova, M, Artamonova, T, Khodorkovskii, M, Konevega, A.L, Sergiev, P.V, Severinov, K, Polikanov, Y.S. | | Deposit date: | 2017-06-12 | | Release date: | 2017-08-30 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Klebsazolicin inhibits 70S ribosome by obstructing the peptide exit tunnel.
Nat. Chem. Biol., 13, 2017
|
|
7D0J
 
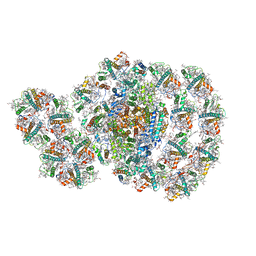 | | Photosystem I-LHCI-LHCII of Chlamydomonas reinhardtii | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Wang, W.D, Shen, L.L, Huang, Z.H, Han, G.Y, Zhang, X, Shen, J.R. | | Deposit date: | 2020-09-10 | | Release date: | 2021-03-03 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | Structure of photosystem I-LHCI-LHCII from the green alga Chlamydomonas reinhardtii in State 2.
Nat Commun, 12, 2021
|
|
7A4H
 
 | |
7A4G
 
 | |
7N2V
 
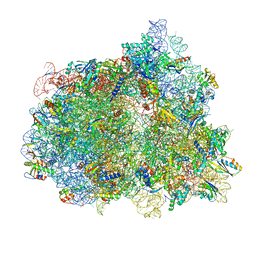 | | Elongating 70S ribosome complex in a spectinomycin-stalled intermediate state of translocation bound to EF-G in an active, GTP conformation (INT1) | | Descriptor: | 1,4-DIAMINOBUTANE, 16S rRNA, 23S rRNA, ... | | Authors: | Rundlet, E.J, Holm, M, Schacherl, M, Natchiar, K.S, Altman, R.B, Spahn, C.M.T, Myasnikov, A.G, Blanchard, S.C. | | Deposit date: | 2021-05-29 | | Release date: | 2021-07-14 | | Last modified: | 2021-08-11 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | Structural basis of early translocation events on the ribosome.
Nature, 595, 2021
|
|
7AC7
 
 | | Structure of accomodated trans-translation complex on E. Coli stalled ribosome. | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Guyomar, C, D'Urso, G, Chat, S, Giudice, E, Gillet, R. | | Deposit date: | 2020-09-10 | | Release date: | 2021-08-18 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Structures of tmRNA and SmpB as they transit through the ribosome.
Nat Commun, 12, 2021
|
|
7A0S
 
 | | 50S Deinococcus radiodurans ribosome bounded with mycinamicin I | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Breiner, E, Eyal, Z, Matzov, D, Halfon, Y, Cimicata, G, Rozenberg, H, Zimmerman, E, Bashan, A, Yonath, A. | | Deposit date: | 2020-08-10 | | Release date: | 2021-08-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | Ribosome-binding and anti-microbial studies of the mycinamicins, 16-membered macrolide antibiotics from Micromonospora griseorubida.
Nucleic Acids Res., 49, 2021
|
|
7A0R
 
 | | 50S Deinococcus radiodurans ribosome bounded with mycinamicin I | | Descriptor: | 50S ribosomal protein L13, 50S ribosomal protein L14, 50S ribosomal protein L15, ... | | Authors: | Breiner, E, Eyal, Z, Matzov, D, Halfon, Y, Cimicata, G, Rozenberg, H, Zimmerman, E, Bashan, A, Yonath, A. | | Deposit date: | 2020-08-10 | | Release date: | 2021-08-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Ribosome-binding and anti-microbial studies of the mycinamicins, 16-membered macrolide antibiotics from Micromonospora griseorubida.
Nucleic Acids Res., 49, 2021
|
|
7NBU
 
 | | Structure of the HigB1 toxin mutant K95A from Mycobacterium tuberculosis (Rv1955) and its target, the cspA mRNA, on the E. coli Ribosome. | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Giudice, E, Mansour, M, Chat, S, D'Urso, G, Gillet, R, Genevaux, P. | | Deposit date: | 2021-01-27 | | Release date: | 2022-03-02 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Substrate recognition and cryo-EM structure of the ribosome-bound TAC toxin of Mycobacterium tuberculosis.
Nat Commun, 13, 2022
|
|
7DR2
 
 | | Structure of GraFix PSI tetramer from Cyanophora paradoxa | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Kato, K, Nagao, R, Akita, F, Miyazaki, N, Shen, J.R. | | Deposit date: | 2020-12-25 | | Release date: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural insights into an evolutionary turning-point of photosystem I from prokaryotes to eukaryotes
Biorxiv, 2022
|
|
7NPA
 
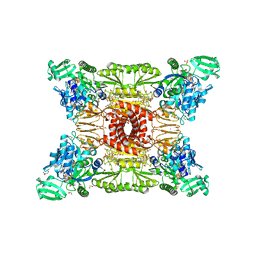 | |
7N8B
 
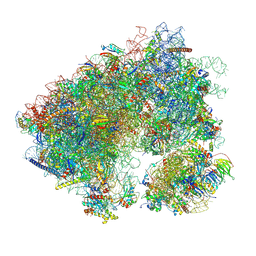 | | Cycloheximide bound vacant 80S structure isolated from cbf5-D95A | | Descriptor: | 18S RIBOSOMAL RNA, 25S, 4-{(2R)-2-[(1S,3S,5S)-3,5-dimethyl-2-oxocyclohexyl]-2-hydroxyethyl}piperidine-2,6-dione, ... | | Authors: | Rai, J, Zhao, Y, Li, H. | | Deposit date: | 2021-06-14 | | Release date: | 2022-05-11 | | Last modified: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | CryoEM structures of pseudouridine-free ribosome suggest impacts of chemical modifications on ribosome conformations.
Structure, 30, 2022
|
|
5JC9
 
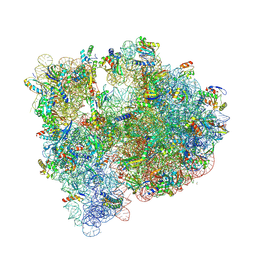 | | Structure of the Escherichia coli ribosome with the U1052G mutation in the 16S rRNA | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 1,4-DIAMINOBUTANE, ... | | Authors: | Cocozaki, A, Ferguson, A. | | Deposit date: | 2016-04-14 | | Release date: | 2016-07-06 | | Last modified: | 2016-08-03 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | Resistance mutations generate divergent antibiotic susceptibility profiles against translation inhibitors.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5J7L
 
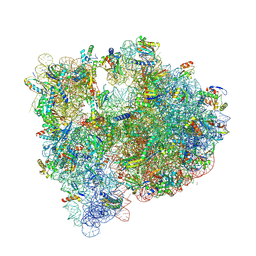 | | Structure of the 70S E coli ribosome with the U1052G mutation in the 16S rRNA bound to tetracycline | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 1,4-DIAMINOBUTANE, ... | | Authors: | Cocozaki, A, Ferguson, A. | | Deposit date: | 2016-04-06 | | Release date: | 2016-07-27 | | Last modified: | 2018-08-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Resistance mutations generate divergent antibiotic susceptibility profiles against translation inhibitors.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5IBB
 
 | | Structure of T. thermophilus 70S ribosome complex with mRNA, tRNAfMet and cognate tRNAVal in the A-site | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Rozov, A, Demeshkina, N, Yusupov, M, Yusupova, G. | | Deposit date: | 2016-02-22 | | Release date: | 2016-05-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | The ribosome prohibits the GU wobble geometry at the first position of the codon-anticodon helix.
Nucleic Acids Res., 44, 2016
|
|
5IB8
 
 | | Structure of T. thermophilus 70S ribosome complex with mRNA, tRNAfMet and near-cognate tRNALys with U-G mismatch in the A-site | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Rozov, A, Demeshkina, N, Yusupov, M, Yusupova, G. | | Deposit date: | 2016-02-22 | | Release date: | 2016-05-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.13 Å) | | Cite: | The ribosome prohibits the GU wobble geometry at the first position of the codon-anticodon helix.
Nucleic Acids Res., 44, 2016
|
|
7B5K
 
 | | E. coli 70S containing suppressor tRNA in the A-site stabilized by a Negamycin analogue and P-site tRNA-nascent chain. | | Descriptor: | 16S rRNA, 2-[[[(3~{R},5~{R})-3-azanyl-6-(3-azanylpropylamino)-5-oxidanyl-hexanoyl]amino]-methyl-amino]ethanoic acid, 23S rRNA, ... | | Authors: | Albers, S, Beckert, B, Matthies, M, Schuster, R, Riedner, M, Seuring, C, Sanyal, S, Torda, A, Wilson, N.D, Ignatova, Z. | | Deposit date: | 2020-12-03 | | Release date: | 2021-05-26 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Harnessing evolutionarily flexibility in tRNAs to maximize nonsense suppression activities
Nat Commun, 2021
|
|
5J3C
 
 | |
5J88
 
 | | Structure of the E coli 70S ribosome with the U1060A mutation in 16S rRNA | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 1,4-DIAMINOBUTANE, ... | | Authors: | Cocozaki, A, Ferguson, A. | | Deposit date: | 2016-04-07 | | Release date: | 2016-07-06 | | Last modified: | 2016-12-07 | | Method: | X-RAY DIFFRACTION (3.32 Å) | | Cite: | Resistance mutations generate divergent antibiotic susceptibility profiles against translation inhibitors.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
7O0W
 
 | | Cryo-EM structure of the RC-dLH complex (model_1b) from Gemmatimonas phototrophica at 2.47 A | | Descriptor: | (19R,22S)-25-amino-22-hydroxy-22-oxido-16-oxo-17,21,23-trioxa-22lambda~5~-phosphapentacosan-19-yl (9Z)-hexadec-9-enoate, (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, (2R,5R,11R,14R)-5,8,11-trihydroxy-5,11-dioxido-17-oxo-2,14-bis(tetradecanoyloxy)-4,6,10,12,16-pentaoxa-5,11-diphosphatriacont-1-yl tetradecanoate, ... | | Authors: | Qian, P, Koblizek, M. | | Deposit date: | 2021-03-27 | | Release date: | 2022-03-02 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (2.47 Å) | | Cite: | 2.4- angstrom structure of the double-ring Gemmatimonas phototrophica photosystem.
Sci Adv, 8, 2022
|
|
7O0U
 
 | | Cryo-EM structure (model_1a) of the RC-dLH complex from Gemmatimonas phototrophica at 2.4 A | | Descriptor: | (19R,22S)-25-amino-22-hydroxy-22-oxido-16-oxo-17,21,23-trioxa-22lambda~5~-phosphapentacosan-19-yl (9Z)-hexadec-9-enoate, (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, (2R,5R,11R,14R)-5,8,11-trihydroxy-5,11-dioxido-17-oxo-2,14-bis(tetradecanoyloxy)-4,6,10,12,16-pentaoxa-5,11-diphosphatriacont-1-yl tetradecanoate, ... | | Authors: | Qian, P, Koblizek, M. | | Deposit date: | 2021-03-27 | | Release date: | 2022-03-02 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (2.35 Å) | | Cite: | 2.4- angstrom structure of the double-ring Gemmatimonas phototrophica photosystem.
Sci Adv, 8, 2022
|
|
7O0X
 
 | | Cryo-EM structure (model_2b) of the RC-dLH complex from Gemmatimonas phototrophica at 2.44 A | | Descriptor: | (19R,22S)-25-amino-22-hydroxy-22-oxido-16-oxo-17,21,23-trioxa-22lambda~5~-phosphapentacosan-19-yl (9Z)-hexadec-9-enoate, (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, (2R,5R,11R,14R)-5,8,11-trihydroxy-5,11-dioxido-17-oxo-2,14-bis(tetradecanoyloxy)-4,6,10,12,16-pentaoxa-5,11-diphosphatriacont-1-yl tetradecanoate, ... | | Authors: | Qian, P, Koblizek, M. | | Deposit date: | 2021-03-28 | | Release date: | 2022-03-02 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (2.44 Å) | | Cite: | 2.4- angstrom structure of the double-ring Gemmatimonas phototrophica photosystem.
Sci Adv, 8, 2022
|
|
7O0V
 
 | | Cryo-EM structure (model_2a) of the RC-dLH complex from Gemmatimonas phototrophica at 2.5 A | | Descriptor: | (19R,22S)-25-amino-22-hydroxy-22-oxido-16-oxo-17,21,23-trioxa-22lambda~5~-phosphapentacosan-19-yl (9Z)-hexadec-9-enoate, (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, (2R,5R,11R,14R)-5,8,11-trihydroxy-5,11-dioxido-17-oxo-2,14-bis(tetradecanoyloxy)-4,6,10,12,16-pentaoxa-5,11-diphosphatriacont-1-yl tetradecanoate, ... | | Authors: | Qian, P, Koblizek, M. | | Deposit date: | 2021-03-27 | | Release date: | 2022-03-02 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | 2.4- angstrom structure of the double-ring Gemmatimonas phototrophica photosystem.
Sci Adv, 8, 2022
|
|
7EXT
 
 | | Cryo-EM structure of cyanobacterial phycobilisome from Synechococcus sp. PCC 7002 | | Descriptor: | Allophycocyanin alpha subunit, Allophycocyanin beta subunit, Allophycocyanin subunit alpha-B, ... | | Authors: | Zheng, L, Zheng, Z, Li, X, Wang, G, Zhang, K, Wei, P, Zhao, J, Gao, N. | | Deposit date: | 2021-05-28 | | Release date: | 2021-10-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural insight into the mechanism of energy transfer in cyanobacterial phycobilisomes.
Nat Commun, 12, 2021
|
|
7EYD
 
 | | Cryo-EM structure of cyanobacterial phycobilisome from Anabaena sp. PCC 7120 | | Descriptor: | Allophycocyanin subunit alpha 1, Allophycocyanin subunit alpha-B, Allophycocyanin subunit beta, ... | | Authors: | Zheng, L, Zheng, Z, Li, X, Wang, G, Zhang, K, Wei, P, Zhao, J, Gao, N. | | Deposit date: | 2021-05-30 | | Release date: | 2021-10-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural insight into the mechanism of energy transfer in cyanobacterial phycobilisomes.
Nat Commun, 12, 2021
|
|
