1HMK
 
 | | RECOMBINANT GOAT ALPHA-LACTALBUMIN | | Descriptor: | CALCIUM ION, PROTEIN (ALPHA-LACTALBUMIN) | | Authors: | Horii, K, Matsushima, M, Tsumoto, K, Kumagai, I. | | Deposit date: | 1998-11-26 | | Release date: | 1999-11-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Effect of the extra n-terminal methionine residue on the stability and folding of recombinant alpha-lactalbumin expressed in Escherichia coli.
J.Mol.Biol., 285, 1999
|
|
2EXJ
 
 | | Structure of the family43 beta-Xylosidase D128G mutant from geobacillus stearothermophilus in complex with xylobiose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GLYCEROL, ... | | Authors: | Brux, C, Niefind, K, Shallom-Shezifi, D, Shoham, Y, Schomburg, D. | | Deposit date: | 2005-11-08 | | Release date: | 2006-04-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Structure of an Inverting GH43 beta-Xylosidase from Geobacillus stearothermophilus with its Substrate Reveals the Role of the Three Catalytic Residues.
J.Mol.Biol., 359, 2006
|
|
1ZIN
 
 | | ADENYLATE KINASE WITH BOUND AP5A | | Descriptor: | ADENYLATE KINASE, BIS(ADENOSINE)-5'-PENTAPHOSPHATE, ZINC ION | | Authors: | Berry, M.B, Phillips Jr, G.N. | | Deposit date: | 1996-06-07 | | Release date: | 1997-06-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of Bacillus stearothermophilus adenylate kinase with bound Ap5A, Mg2+ Ap5A, and Mn2+ Ap5A reveal an intermediate lid position and six coordinate octahedral geometry for bound Mg2+ and Mn2+.
Proteins, 32, 1998
|
|
2EWT
 
 | | Crystal structure of the DNA-binding domain of BldD | | Descriptor: | SULFATE ION, putative DNA-binding protein | | Authors: | Kim, I.K, Lee, C.J, Kim, M.K, Kim, J.M, Kim, J.H, Yim, H.S, Cha, S.S, Kang, S.O. | | Deposit date: | 2005-11-07 | | Release date: | 2006-06-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystal structure of the DNA-binding domain of BldD, a central regulator of aerial mycelium formation in Streptomyces coelicolor A3(2)
Mol.Microbiol., 60, 2006
|
|
1HP7
 
 | | A 2.1 ANGSTROM STRUCTURE OF AN UNCLEAVED ALPHA-1-ANTITRYPSIN SHOWS VARIABILITY OF THE REACTIVE CENTER AND OTHER LOOPS | | Descriptor: | ALPHA-1-ANTITRYPSIN, BETA-MERCAPTOETHANOL, ZINC ION | | Authors: | Kim, S.-J, Woo, J.-R, Seo, E.J, Yu, M.-H, Ryu, S.-E. | | Deposit date: | 2000-12-12 | | Release date: | 2001-03-14 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A 2.1 A resolution structure of an uncleaved alpha(1)-antitrypsin shows variability of the reactive center and other loops.
J.Mol.Biol., 306, 2001
|
|
2EXK
 
 | | Structure of the family43 beta-Xylosidase E187G from geobacillus stearothermophilus in complex with xylobiose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GLYCEROL, ... | | Authors: | Brux, C, Niefind, K, Shallom-Shezifi, D, Shoham, Y, Schomburg, D. | | Deposit date: | 2005-11-08 | | Release date: | 2006-04-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Structure of an Inverting GH43 beta-Xylosidase from Geobacillus stearothermophilus with its Substrate Reveals the Role of the Three Catalytic Residues.
J.Mol.Biol., 359, 2006
|
|
1XOF
 
 | | Heterooligomeric Beta Beta Alpha Miniprotein | | Descriptor: | BBAhetT1 | | Authors: | Ali, M.H, Taylor, C.M, Grigoryan, G, Allen, K.N, Imperiali, B, Keating, A.E. | | Deposit date: | 2004-10-06 | | Release date: | 2005-02-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Design of a Heterospecific, Tetrameric, 21-Residue Miniprotein with Mixed alpha/beta Structure.
Structure, 13, 2005
|
|
4A7P
 
 | | Se-Met derivatized UgdG, UDP-glucose dehydrogenase from Sphingomonas elodea | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, UDP-GLUCOSE DEHYDROGENASE | | Authors: | Rocha, J, Granja, A.T, Sa-Correia, I, Fialho, A.M, Frazao, C. | | Deposit date: | 2011-11-14 | | Release date: | 2012-11-21 | | Last modified: | 2019-09-25 | | Method: | X-RAY DIFFRACTION (3.403 Å) | | Cite: | The Structure of Ugdg, an Udp-Glucose Dehydrogenase from Sphingomonas Elodea Atcc 31461
To be Published
|
|
1BKH
 
 | | MUCONATE LACTONIZING ENZYME FROM PSEUDOMONAS PUTIDA | | Descriptor: | MUCONATE LACTONIZING ENZYME | | Authors: | Hasson, M.S, Schlichting, I, Moulai, J, Taylor, K, Barrett, W, Kenyon, G.L, Babbitt, P.C, Gerlt, J.A, Petsko, G.A, Ringe, D. | | Deposit date: | 1998-07-07 | | Release date: | 1998-10-21 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Evolution of an enzyme active site: the structure of a new crystal form of muconate lactonizing enzyme compared with mandelate racemase and enolase.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1GN9
 
 | | Hybrid Cluster Protein from Desulfovibrio desulfuricans ATCC 27774 X-ray structure at 2.6A resolution using synchrotron radiation at a wavelength of 1.722A | | Descriptor: | HYBRID CLUSTER PROTEIN, IRON/SULFUR CLUSTER, IRON/SULFUR/OXYGEN HYBRID CLUSTER | | Authors: | Macedo, S, Mitchell, E.P, Romao, C.V, Cooper, S.J, Coelho, R, Liu, M.Y, Xavier, A.V, Legall, J, Bailey, S, Garner, D.C, Hagen, W.R, Teixeira, M, Carrondo, M.A, Lindley, P. | | Deposit date: | 2001-10-04 | | Release date: | 2002-04-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Hybrid Cluster Proteins (Hcps) from Desulfovibrio Desulfuricans Atcc 27774 and Desulfovibrio Vulgaris (Hildenborough): X-Ray Structures at 1.25 A Resolution Using Synchrotron Radiation.
J.Biol.Inorg.Chem., 7, 2002
|
|
4ABN
 
 | | Crystal structure of full length mouse Strap (TTC5) | | Descriptor: | 1,2-ETHANEDIOL, TETRATRICOPEPTIDE REPEAT PROTEIN 5 | | Authors: | Pike, A.C.W, Bullock, A.N, Kleinekofort, W, Zimmermann, T, Burgess-Brown, N, Sharpe, T.D, Thangaratnarajah, C, Keates, T, Ugochukwu, E, Bunkoczi, G, Uppenberg, J, von Delft, F, Arrowsmith, C.H, Weigelt, J, Edwards, A, Bountra, C, La Thangue, N.B, Knapp, S. | | Deposit date: | 2011-12-09 | | Release date: | 2012-01-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The P53 Cofactor Strap Exhibits an Unexpected Tpr Motif and Oligonucleotide-Binding (Ob)-Fold Structure.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1GAB
 
 | | STRUCTURE OF AN ALBUMIN-BINDING DOMAIN, NMR, 20 STRUCTURES | | Descriptor: | PROTEIN PAB | | Authors: | Johansson, M.U, De Chateau, M, Wikstrom, M, Forsen, S, Drakenberg, T, Bjorck, L. | | Deposit date: | 1996-12-30 | | Release date: | 1997-07-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the albumin-binding GA module: a versatile bacterial protein domain.
J.Mol.Biol., 266, 1997
|
|
1M05
 
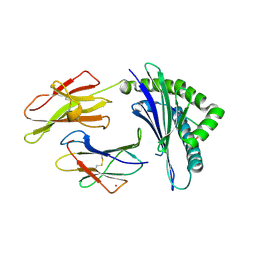 | | HLA B8 in complex with an Epstein Barr Virus determinant | | Descriptor: | Beta-2-microglobulin, CADMIUM ION, EBNA-3 nuclear protein, ... | | Authors: | Kjer-Nielsen, L, Clements, C.S, Brooks, A.G, Purcell, A.W, Fontes, M.R, McCluskey, J, Rossjohn, J. | | Deposit date: | 2002-06-11 | | Release date: | 2003-09-02 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structure of HLA-B8 complexed to an immunodominant viral determinant: peptide-induced conformational changes and
a mode of MHC class I dimerization.
J.IMMUNOL., 169, 2003
|
|
1D42
 
 | |
1XY1
 
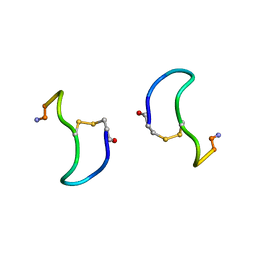 | | CRYSTAL STRUCTURE ANALYSIS OF DEAMINO-OXYTOCIN. CONFORMATIONAL FLEXIBILITY AND RECEPTOR BINDING | | Descriptor: | BETA-MERCAPTOPROPIONATE-OXYTOCIN | | Authors: | Husain, J, Blundell, T.L, Wood, S.P, Tickle, I.J, Cooper, S, Pitts, J.E. | | Deposit date: | 1987-06-05 | | Release date: | 1988-04-16 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Crystal structure analysis of deamino-oxytocin: conformational flexibility and receptor binding.
Science, 232, 1986
|
|
1XUE
 
 | |
2ERQ
 
 | | Crystal structure of vascular apoptosis-inducing protein-1(tetragonal crystal form) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Takeda, S, Igarashi, T, Araki, S. | | Deposit date: | 2005-10-25 | | Release date: | 2006-06-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of VAP1 reveal ADAMs' MDC domain architecture and its unique C-shaped scaffold
Embo J., 25, 2006
|
|
2O81
 
 | |
2O83
 
 | |
1Z37
 
 | | Crystal structure of Trichomonas vaginalis purine nucleoside phosphorylase complexed with adenosine | | Descriptor: | ADENOSINE, purine nucleoside phosphorylase | | Authors: | Zhang, Y, Wang, W.H, Wu, S.W, Wang, C.C, Ealick, S.E. | | Deposit date: | 2005-03-10 | | Release date: | 2005-03-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Identification of a subversive substrate of Trichomonas vaginalis purine nucleoside phosphorylase and the crystal structure of the enzyme-substrate complex.
J.Biol.Chem., 280, 2005
|
|
1IBD
 
 | | X-RAY 3D STRUCTURE OF P.LEIOGNATHI CU,ZN SOD MUTANT V29A | | Descriptor: | COPPER (II) ION, CU,ZN SUPEROXIDE DISMUTASE, ZINC ION | | Authors: | Stroppolo, M.E, Pesce, A, D'Orazio, M, O'Neill, P, Bordo, D, Rosano, C, Milani, M, Battistoni, A, Bolognesi, M, Desideri, A. | | Deposit date: | 2001-03-28 | | Release date: | 2001-05-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Single mutations at the subunit interface modulate copper reactivity in Photobacterium leiognathi Cu,Zn superoxide dismutase.
J.Mol.Biol., 308, 2001
|
|
2ADM
 
 | | ADENINE-N6-DNA-METHYLTRANSFERASE TAQI | | Descriptor: | ADENINE-N6-DNA-METHYLTRANSFERASE TAQI, S-ADENOSYLMETHIONINE | | Authors: | Schluckebier, G, Saenger, W. | | Deposit date: | 1996-07-15 | | Release date: | 1997-01-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Differential binding of S-adenosylmethionine S-adenosylhomocysteine and Sinefungin to the adenine-specific DNA methyltransferase M.TaqI.
J.Mol.Biol., 265, 1997
|
|
2AAA
 
 | | CALCIUM BINDING IN ALPHA-AMYLASES: AN X-RAY DIFFRACTION STUDY AT 2.1 ANGSTROMS RESOLUTION OF TWO ENZYMES FROM ASPERGILLUS | | Descriptor: | ALPHA-AMYLASE, CALCIUM ION | | Authors: | Brady, L, Brzozowski, A.M, Derewenda, Z, Dodson, E.J, Dodson, G.G. | | Deposit date: | 1991-02-27 | | Release date: | 1993-07-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Calcium binding in alpha-amylases: an X-ray diffraction study at 2.1-A resolution of two enzymes from Aspergillus.
Biochemistry, 29, 1990
|
|
1IB5
 
 | | X-RAY 3D STRUCTURE OF P.LEIOGNATHI CU,ZN SOD MUTANT W83Y | | Descriptor: | COPPER (II) ION, CU,ZN SUPEROXIDE DISMUTASE, ZINC ION | | Authors: | Stroppolo, M.E, Pesce, A, D'Orazio, M, O'Neill, P, Bordo, D, Rosano, C, Milani, M, Battistoni, A, Bolognesi, M, Desideri, A. | | Deposit date: | 2001-03-27 | | Release date: | 2001-05-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Single mutations at the subunit interface modulate copper reactivity in Photobacterium leiognathi Cu,Zn superoxide dismutase.
J.Mol.Biol., 308, 2001
|
|
1C1C
 
 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH TNK-6123 | | Descriptor: | 6-(cyclohexylsulfanyl)-1-(ethoxymethyl)-5-(1-methylethyl)pyrimidine-2,4(1H,3H)-dione, HIV-1 REVERSE TRANSCRIPTASE (A-CHAIN), HIV-1 REVERSE TRANSCRIPTASE (B-CHAIN) | | Authors: | Hopkins, A.L, Ren, J, Tanaka, H, Baba, M, Okamato, M, Stuart, D.I, Stammers, D.K. | | Deposit date: | 1999-07-21 | | Release date: | 2000-07-21 | | Last modified: | 2014-11-12 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Design of MKC-442 (emivirine) analogues with improved activity against drug-resistant HIV mutants.
J.Med.Chem., 42, 1999
|
|
