1PIV
 
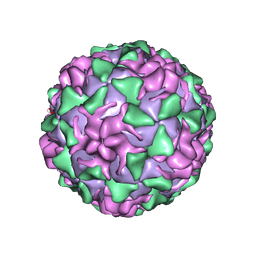 | | BINDING OF THE ANTIVIRAL DRUG WIN51711 TO THE SABIN STRAIN OF TYPE 3 POLIOVIRUS: STRUCTURAL COMPARISON WITH DRUG BINDING IN RHINOVIRUS 14 | | Descriptor: | 5-(7-(4-(4,5-DIHYDRO-2-OXAZOLYL)PHENOXY)HEPTYL)-3-METHYL ISOXAZOLE, MYRISTIC ACID, POLIOVIRUS TYPE 3 (SUBUNIT VP1), ... | | Authors: | Hiremath, C.N, Grant, R.A, Filman, D.J, Hogle, J.M. | | Deposit date: | 1995-02-02 | | Release date: | 1995-06-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Binding of the antiviral drug WIN51711 to the sabin strain of type 3 poliovirus: structural comparison with drug binding in rhinovirus 14.
Acta Crystallogr.,Sect.D, 51, 1995
|
|
1PX6
 
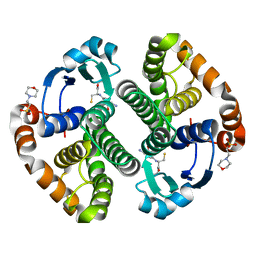 | | A folding mutant of human class pi glutathione transferase, created by mutating aspartate 153 of the wild-type protein to asparagine | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLUTATHIONE, Glutathione S-transferase P | | Authors: | Kong, G.K.-W, Polekhina, G, McKinstry, W.J, Parker, M.W, Dragani, B, Aceto, A, Paludi, D, Principe, D.R, Mannervik, B, Stenberg, G. | | Deposit date: | 2003-07-02 | | Release date: | 2003-07-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The multi-functional role of a highly conserved aspartic acid residue in glutathione transferase P1-1
To be Published
|
|
1PXS
 
 | | Structure of Met56Ala mutant of Bacteriorhodopsin | | Descriptor: | Bacteriorhodopsin, RETINAL | | Authors: | Faham, S, Yang, D, Bare, E, Yohannan, S, Whitelegge, J.P, Bowie, J.U. | | Deposit date: | 2003-07-06 | | Release date: | 2003-12-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Side-chain Contributions to Membrane Protein Structure and Stability.
J.Mol.Biol., 335, 2004
|
|
1PYA
 
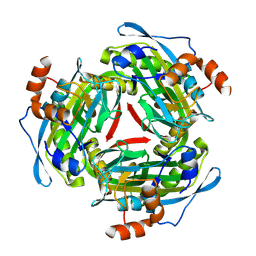 | |
1POC
 
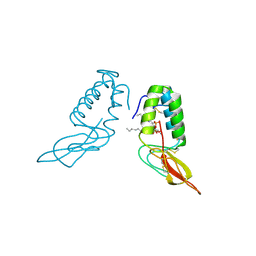 | |
1PPL
 
 | |
1PWQ
 
 | | Crystal structure of Anthrax Lethal Factor complexed with Thioacetyl-Tyr-Pro-Met-Amide, a metal-chelating peptidyl small molecule inhibitor | | Descriptor: | Lethal factor, N-(SULFANYLACETYL)TYROSYLPROLYLMETHIONINAMIDE, ZINC ION | | Authors: | Wong, T.Y, Schwarzenbacher, R, Liddington, R.C. | | Deposit date: | 2003-07-02 | | Release date: | 2004-01-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.52 Å) | | Cite: | The structural basis for substrate and inhibitor selectivity of the anthrax lethal factor.
Nat.Struct.Mol.Biol., 11, 2004
|
|
1PLU
 
 | |
1PPC
 
 | | GEOMETRY OF BINDING OF THE BENZAMIDINE-AND ARGININE-BASED INHIBITORS N-ALPHA-(2-NAPHTHYL-SULPHONYL-GLYCYL)-DL-P-AMIDINOPHENYLALANYL-PIPERIDINE (NAPAP) AND (2R,4R)-4-METHYL-1-[N-ALPHA-(3-METHYL-1,2,3,4-TETRAHYDRO-8-QUINOLINESULPHONYL)-L-ARGINYL]-2-PIPERIDINE CARBOXYLIC ACID (MQPA) TO HUMAN ALPHA-THROMBIN: X-RAY CRYSTALLOGRAPHIC DETERMINATION OF THE NAPAP-TRYPSIN COMPLEX AND MODELING OF NAPAP-THROMBIN AND MQPA-THROMBIN | | Descriptor: | 1-[N-(naphthalen-2-ylsulfonyl)glycyl-4-carbamimidoyl-D-phenylalanyl]piperidine, CALCIUM ION, TRYPSIN | | Authors: | Bode, W, Turk, D. | | Deposit date: | 1991-10-24 | | Release date: | 1994-01-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Geometry of binding of the benzamidine- and arginine-based inhibitors N alpha-(2-naphthyl-sulphonyl-glycyl)-DL-p-amidinophenylalanyl-pipe ridine (NAPAP) and (2R,4R)-4-methyl-1-[N alpha-(3-methyl-1,2,3,4-tetrahydr quinolinesulphonyl)-L-arginyl]-2-piperidine carboxylic acid (MQPA) to human alpha-thrombin.X-ray crystallographic determination of the NAPAP-trypsin complex and modeling of NAPAP-thrombin and MQPA-thrombin.
Eur.J.Biochem., 193, 1990
|
|
1PO0
 
 | |
1POO
 
 | | THERMOSTABLE PHYTASE FROM BACILLUS SP | | Descriptor: | CALCIUM ION, PROTEIN (PHYTASE) | | Authors: | Oh, B.H, Ha, N.C. | | Deposit date: | 1999-04-16 | | Release date: | 2000-04-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of a novel, thermostable phytase in partially and fully calcium-loaded states.
Nat.Struct.Biol., 7, 2000
|
|
1Q0X
 
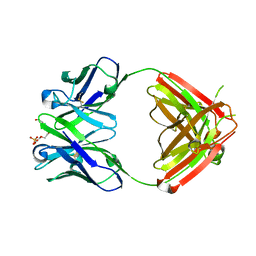 | | Anti-morphine Antibody 9B1 Unliganded Form | | Descriptor: | Fab 9B1, heavy chain, light chain, ... | | Authors: | Pozharski, E, Wilson, M.A, Hewagama, A, Shanafelt, A.B, Petsko, G, Ringe, D. | | Deposit date: | 2003-07-17 | | Release date: | 2004-04-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Anchoring a cationic ligand: the structure of the Fab fragment of the anti-morphine antibody 9B1 and its complex with morphine
J.Mol.Biol., 337, 2004
|
|
1POH
 
 | |
1POP
 
 | |
1PVI
 
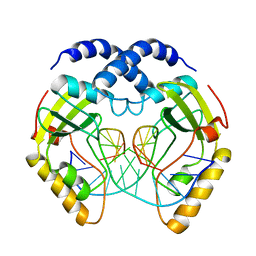 | | STRUCTURE OF PVUII ENDONUCLEASE WITH COGNATE DNA | | Descriptor: | DNA (5'-D(*TP*GP*AP*CP*CP*AP*GP*CP*TP*GP*GP*TP*C)-3'), PROTEIN (PVUII (E.C.3.1.21.4)) | | Authors: | Cheng, X, Balendiran, K, Schildkraut, I, Anderson, J.E. | | Deposit date: | 1994-11-16 | | Release date: | 1995-02-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of PvuII endonuclease with cognate DNA.
EMBO J., 13, 1994
|
|
1PWW
 
 | |
1Q06
 
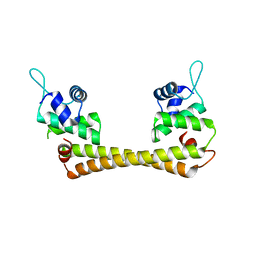 | | Crystal structure of the Ag(I) form of E. coli CueR, a copper efflux regulator | | Descriptor: | SILVER ION, Transcriptional regulator cueR | | Authors: | Changela, A, Chen, K, Xue, Y, Holschen, J, Outten, C.E, O'Halloran, T.V, Mondragon, A. | | Deposit date: | 2003-07-15 | | Release date: | 2003-09-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Molecular basis of metal-ion selectivity and zeptomolar sensitivity by CueR
Science, 301, 2003
|
|
1OD6
 
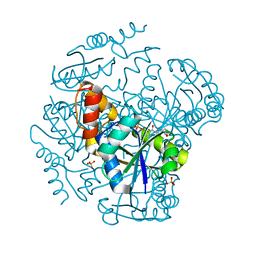 | | The Crystal Structure of Phosphopantetheine adenylyltransferase from Thermus Thermophilus in complex with 4'-phosphopantetheine | | Descriptor: | 4'-PHOSPHOPANTETHEINE, PHOSPHOPANTETHEINE ADENYLYLTRANSFERASE, SULFATE ION | | Authors: | Takahashi, H, Inagaki, E, Miyano, M, Tahirov, T.H. | | Deposit date: | 2003-02-13 | | Release date: | 2003-03-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure and Implications for the Thermal Stability of Phosphopantetheine Adenylyltransferase from Thermus Thermophilus.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1NLT
 
 | | The crystal structure of Hsp40 Ydj1 | | Descriptor: | Mitochondrial protein import protein MAS5, Seven residue peptide, ZINC ION | | Authors: | Li, J, Sha, B. | | Deposit date: | 2003-01-07 | | Release date: | 2004-01-13 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The crystal structure of the yeast Hsp40 Ydj1 complexed with its peptide substrate.
Structure, 11, 2003
|
|
1NM0
 
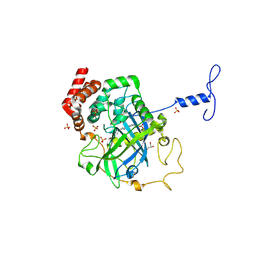 | | Proteus mirabilis catalase in complex with formiate | | Descriptor: | Catalase, FORMIC ACID, GLYCEROL, ... | | Authors: | Andreoletti, P, Pernoud, A, Gouet, P, Jouve, H.M. | | Deposit date: | 2003-01-08 | | Release date: | 2004-01-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural studies of Proteus mirabilis catalase in its ground state, oxidized state and in complex with formic acid.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1NMT
 
 | |
1NSI
 
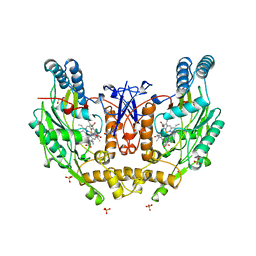 | | HUMAN INDUCIBLE NITRIC OXIDE SYNTHASE, ZN-BOUND, L-ARG COMPLEX | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ARGININE, GLYCEROL, ... | | Authors: | Li, H, Raman, C.S, Glaser, C.B, Blasko, E, Young, T.A, Parkinson, J.F, Whitlow, M, Poulos, T.L. | | Deposit date: | 1999-01-10 | | Release date: | 2000-01-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structures of zinc-free and -bound heme domain of human inducible nitric-oxide synthase. Implications for dimer stability and comparison with endothelial nitric-oxide synthase.
J.Biol.Chem., 274, 1999
|
|
1NOY
 
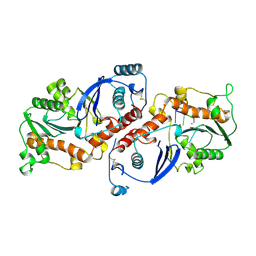 | | DNA POLYMERASE (E.C.2.7.7.7)/DNA COMPLEX | | Descriptor: | DNA (5'-D(*TP*TP*T)-3'), MANGANESE (II) ION, PROTEIN (DNA POLYMERASE (E.C.2.7.7.7)), ... | | Authors: | Wang, J, Yu, P, Lin, T.C, Konigsberg, W.H, Steitz, T.A. | | Deposit date: | 1996-02-16 | | Release date: | 1996-10-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of an NH2-terminal fragment of T4 DNA polymerase and its complexes with single-stranded DNA and with divalent metal ions.
Biochemistry, 35, 1996
|
|
4E7V
 
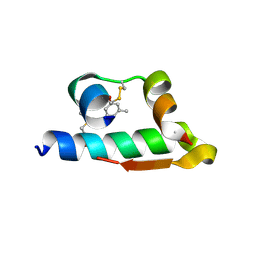 | | The structure of R6 bovine insulin | | Descriptor: | CHLORIDE ION, Insulin A chain, Insulin B chain, ... | | Authors: | Harris, P, Frankaer, C.G, Knudsen, M.V. | | Deposit date: | 2012-03-19 | | Release date: | 2012-04-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structures of T(6), T(3)R(3) and R(6) bovine insulin: combining X-ray diffraction and absorption spectroscopy.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4QI7
 
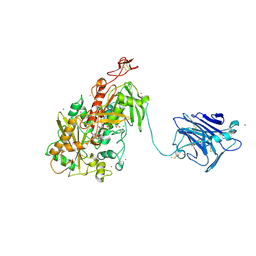 | | Cellobiose dehydrogenase from Neurospora crassa, NcCDH | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cellobiose dehydrogenase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Tan, T.C, Gandini, R, Sygmund, C, Kittl, R, Haltrich, D, Ludwig, R, Hallberg, B.M, Divne, C. | | Deposit date: | 2014-05-30 | | Release date: | 2015-07-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for cellobiose dehydrogenase action during oxidative cellulose degradation.
Nat Commun, 6, 2015
|
|
