6YL4
 
 | | Soluble epoxide hydrolase in complex with 3-((R)-3-(1-hydroxyureido)but-1-yn-1-yl)-N-((S)-3-phenyl-3-(4-trifluoromethoxy)phenyl)propyl)benzamide | | Descriptor: | 3-[(3~{R})-3-[aminocarbonyl(oxidanyl)amino]but-1-ynyl]-~{N}-[(3~{S})-3-phenyl-3-[4-(trifluoromethyloxy)phenyl]propyl]benzamide, Bifunctional epoxide hydrolase 2 | | Authors: | Kramer, J.S, Pogoryelov, D, Hiesinger, K, Proschak, E. | | Deposit date: | 2020-04-06 | | Release date: | 2020-10-21 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Design, Synthesis, and Structure-Activity Relationship Studies of Dual Inhibitors of Soluble Epoxide Hydrolase and 5-Lipoxygenase.
J.Med.Chem., 63, 2020
|
|
6T3G
 
 | |
5H23
 
 | | Crystal structure of Chikungunya virus capsid protein | | Descriptor: | 1,2-ETHANEDIOL, Capsid Protein, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sharma, R, Kesari, P, Tomar, S, Kumar, P. | | Deposit date: | 2016-10-14 | | Release date: | 2018-03-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-function insights into chikungunya virus capsid protein: Small molecules targeting capsid hydrophobic pocket.
Virology, 515, 2018
|
|
9BVA
 
 | | Crystal structure of human CYP3A4 in complex with SJYHJ-106 | | Descriptor: | Cytochrome P450 3A4, N-[2-(2-chloroethyl)phenyl]-N'-[(2P)-2-(1H-imidazol-1-yl)-5-(trifluoromethyl)phenyl]urea, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Jingheng, W, Nithianantham, S, Miller, D.J, Chen, T. | | Deposit date: | 2024-05-20 | | Release date: | 2025-04-16 | | Last modified: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Decoding the selective chemical modulation of CYP3A4.
Nat Commun, 16, 2025
|
|
9FTW
 
 | |
8VCU
 
 | | Crystal structure of the oligomeric rMcL-1 in complex with lactulose | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Hernandez-Santoyo, A, Loera-Rubalcava, J. | | Deposit date: | 2023-12-14 | | Release date: | 2024-12-18 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | A mytilectin from Mytilus californianus: Study of its unique galactoside interactions, oligomerization patterns, and antifungal activity.
Int.J.Biol.Macromol., 308, 2025
|
|
3L57
 
 | |
9BV8
 
 | | Crystal structure of human CYP3A4 in complex with SJYHJ-114 | | Descriptor: | Cytochrome P450 3A4, N-[(2P)-2-(1H-imidazol-1-yl)-5-(trifluoromethyl)phenyl]cyclobutanecarboxamide, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Jingheng, W, Nithianantham, S, Miller, D.J, Chen, T. | | Deposit date: | 2024-05-20 | | Release date: | 2025-04-16 | | Last modified: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Decoding the selective chemical modulation of CYP3A4.
Nat Commun, 16, 2025
|
|
8W2X
 
 | | TAS-120 covalent structure with FGFR2 | | Descriptor: | 1-[(3S)-3-{4-amino-3-[(3,5-dimethoxyphenyl)ethynyl]-1H-pyrazolo[3,4-d]pyrimidin-1-yl}pyrrolidin-1-yl]propan-1-one, Fibroblast growth factor receptor 2, GLYCEROL, ... | | Authors: | Hoffman, I.D, Nelson, K.J, Bensen, D.C, Bailey, J.B. | | Deposit date: | 2024-02-21 | | Release date: | 2025-01-01 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | A model for decoding resistance in precision oncology: acquired resistance to FGFR inhibitors in cholangiocarcinoma.
Ann Oncol, 36, 2025
|
|
9BV9
 
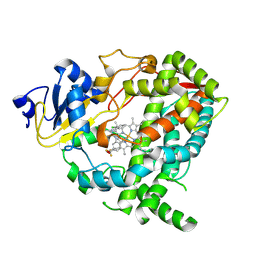 | | Crystal structure of human CYP3A4 in complex with Z56791366 | | Descriptor: | Cytochrome P450 3A4, N-(2-chlorophenyl)-N'-[(2P)-2-(1H-imidazol-1-yl)-5-(trifluoromethyl)phenyl]urea, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Jingheng, W, Nithianantham, S, Miller, D.J, Chen, T. | | Deposit date: | 2024-05-20 | | Release date: | 2025-04-16 | | Last modified: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Decoding the selective chemical modulation of CYP3A4.
Nat Commun, 16, 2025
|
|
8W3B
 
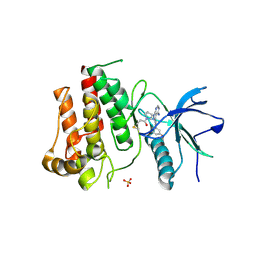 | | TAS-120 covalent structure with FGFR2 molecular brake mutant | | Descriptor: | 1-[(3S)-3-{4-amino-3-[(3,5-dimethoxyphenyl)ethynyl]-1H-pyrazolo[3,4-d]pyrimidin-1-yl}pyrrolidin-1-yl]prop-2-en-1-one, Fibroblast growth factor receptor 2, SULFATE ION | | Authors: | Hoffman, I.D, Nelson, K.J, Bensen, D.C, Bailey, J.B. | | Deposit date: | 2024-02-22 | | Release date: | 2025-01-01 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | A model for decoding resistance in precision oncology: acquired resistance to FGFR inhibitors in cholangiocarcinoma.
Ann Oncol, 36, 2025
|
|
8W3D
 
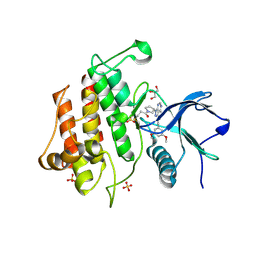 | | TAS-120 covalent structure with FGFR2 molecular brake mutant | | Descriptor: | 1-[(3S)-3-{4-amino-3-[(3,5-dimethoxyphenyl)ethynyl]-1H-pyrazolo[3,4-d]pyrimidin-1-yl}pyrrolidin-1-yl]prop-2-en-1-one, Fibroblast growth factor receptor 2, GLYCEROL, ... | | Authors: | Hoffman, I.D, Nelson, K.J, Bensen, D.C, Bailey, J.B. | | Deposit date: | 2024-02-22 | | Release date: | 2025-01-01 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | A model for decoding resistance in precision oncology: acquired resistance to FGFR inhibitors in cholangiocarcinoma.
Ann Oncol, 36, 2025
|
|
3KN5
 
 | |
7ZTW
 
 | |
8KAE
 
 | | 16d-bound human SPNS2 | | Descriptor: | 3-[3-(4-decylphenyl)-1,2,4-oxadiazol-5-yl]propan-1-amine, NbFab chain L, NbFab-H chain, ... | | Authors: | He, Y, Duan, Y. | | Deposit date: | 2023-08-03 | | Release date: | 2024-01-03 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Structural basis of Sphingosine-1-phosphate transport via human SPNS2.
Cell Res., 34, 2024
|
|
7QRD
 
 | | Crystal structure of mouse CARM1 in complex with histone H3_10-25 | | Descriptor: | (2~{R},3~{R},4~{S},5~{R})-2-(6-aminopurin-9-yl)-5-[(~{E})-prop-1-enyl]oxolane-3,4-diol, 1,2-ETHANEDIOL, Histone-arginine methyltransferase CARM1, ... | | Authors: | Marechal, N, Cura, V, Troffer-Charlier, N, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2022-01-11 | | Release date: | 2022-12-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | CARM1 Transition State Mimics
To Be Published
|
|
7AHI
 
 | | Substrate-engaged type 3 secretion system needle complex from Salmonella enterica typhimurium - SpaR state 2 | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, LAURYL DIMETHYLAMINE-N-OXIDE, Lipoprotein PrgK, ... | | Authors: | Fahrenkamp, D, Goessweiner-Mohr, N, Miletic, S, Wald, J, Marlovits, T. | | Deposit date: | 2020-09-24 | | Release date: | 2021-03-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Substrate-engaged type III secretion system structures reveal gating mechanism for unfolded protein translocation
Nat Commun, 12, 2021
|
|
3KRC
 
 | | Mint heterotetrameric geranyl pyrophosphate synthase in complex with IPP | | Descriptor: | 1,2-ETHANEDIOL, 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, Geranyl diphosphate synthase large subunit, ... | | Authors: | Chang, T.-H, Hsieh, F.-L, Ko, T.-P, Wang, A.H.-J. | | Deposit date: | 2009-11-18 | | Release date: | 2010-02-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of a heterotetrameric geranyl pyrophosphate synthase from mint (Mentha piperita) reveals intersubunit regulation
Plant Cell, 22, 2010
|
|
3KRP
 
 | | Mint heterotetrameric geranyl pyrophosphate synthase in complex with magnesium and GPP | | Descriptor: | 1,2-ETHANEDIOL, GERANYL DIPHOSPHATE, Geranyl diphosphate synthase large subunit, ... | | Authors: | Chang, T.-H, Hsieh, F.-L, Ko, T.-P, Wang, A.H.-J. | | Deposit date: | 2009-11-19 | | Release date: | 2010-02-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structure of a heterotetrameric geranyl pyrophosphate synthase from mint (Mentha piperita) reveals intersubunit regulation
Plant Cell, 22, 2010
|
|
7MAE
 
 | | HIV-1 Protease (I84V) in Complex with PU1 (LR3-46) | | Descriptor: | Protease, SULFATE ION, diethyl [(4-{(2S,3R)-4-{[(4-aminophenyl)sulfonyl][(2S)-2-methylbutyl]amino}-2-[({[(3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl]oxy}carbonyl)amino]-3-hydroxybutyl}phenoxy)methyl]phosphonate | | Authors: | Lockbaum, G.J, Schiffer, C.A. | | Deposit date: | 2021-03-31 | | Release date: | 2022-04-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.735 Å) | | Cite: | HIV-1 Protease in Complex with ligands
To Be Published
|
|
6TAL
 
 | | 3C-like protease from Southampton virus complexed with FMOPL000227a. | | Descriptor: | 5-ethyl-1,3,4-thiadiazol-2-amine, DIMETHYL SULFOXIDE, Genome polyprotein, ... | | Authors: | Guo, J, Cooper, J.B. | | Deposit date: | 2019-10-29 | | Release date: | 2020-08-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | In crystallo-screening for discovery of human norovirus 3C-like protease inhibitors.
J Struct Biol X, 4, 2020
|
|
7QOX
 
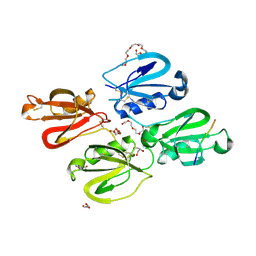 | | Factor XI and Plasma Kallikrein apple domain structures reveals different kininogen bound complexes | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, ... | | Authors: | Li, C, Awital, B, Wong, S, Dreveny, I, Meijers, J, Emsley, J. | | Deposit date: | 2021-12-29 | | Release date: | 2023-01-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Plasma kallikrein structure reveals apple domain disc rotated conformation compared to factor XI.
J Thromb Haemost, 17, 2019
|
|
5N2A
 
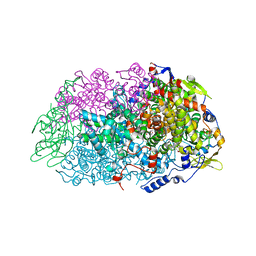 | | METHYL-COENZYME M REDUCTASE III FROM METHANOTORRIS FORMICICUS TRIGONAL FORM | | Descriptor: | 1-THIOETHANESULFONIC ACID, BROMIDE ION, Coenzyme B, ... | | Authors: | Wagner, T, Wegner, C.E, Ermler, U, Shima, S. | | Deposit date: | 2017-02-07 | | Release date: | 2017-06-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Phylogenetic and Structural Comparisons of the Three Types of Methyl Coenzyme M Reductase from Methanococcales and Methanobacteriales.
J.Bacteriol., 199, 2017
|
|
3KQQ
 
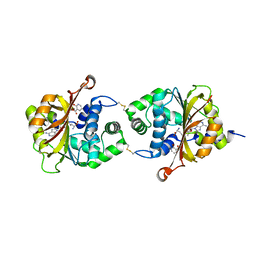 | | Crystal Structure of hPNMT in Complex AdoHcy and 2-Hydroxynicotinic acid | | Descriptor: | 2-oxo-1,2-dihydropyridine-3-carboxylic acid, Phenylethanolamine N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Drinkwater, N, Martin, J.L. | | Deposit date: | 2009-11-17 | | Release date: | 2010-09-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Fragment-based screening by X-ray crystallography, MS and isothermal titration calorimetry to identify PNMT (phenylethanolamine N-methyltransferase) inhibitors.
Biochem.J., 431, 2010
|
|
6W8C
 
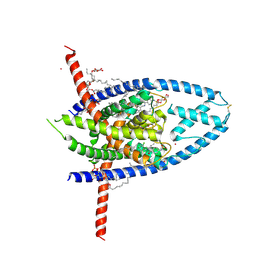 | | K2P2.1 (TREK-1):ML335 complex, 1 mM K+ | | Descriptor: | CADMIUM ION, DODECANE, HEXADECANE, ... | | Authors: | Lolicato, M, Minor, D.L. | | Deposit date: | 2020-03-20 | | Release date: | 2021-01-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | K 2P channel C-type gating involves asymmetric selectivity filter order-disorder transitions.
Sci Adv, 6, 2020
|
|
