1TUB
 
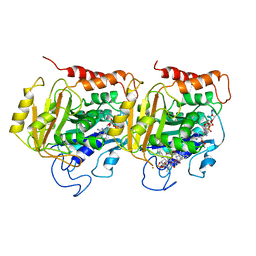 | | TUBULIN ALPHA-BETA DIMER, ELECTRON DIFFRACTION | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, TAXOTERE, ... | | Authors: | Nogales, E, Downing, K.H. | | Deposit date: | 1997-09-23 | | Release date: | 1998-10-07 | | Last modified: | 2024-06-05 | | Method: | ELECTRON CRYSTALLOGRAPHY (3.7 Å) | | Cite: | Structure of the alpha beta tubulin dimer by electron crystallography.
Nature, 391, 1998
|
|
1TUC
 
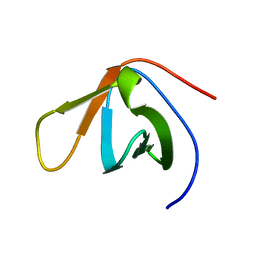 | |
1TUD
 
 | |
1TUE
 
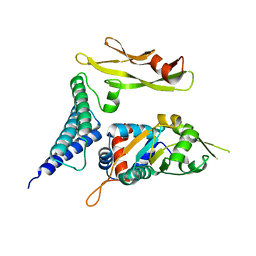 | |
1TUF
 
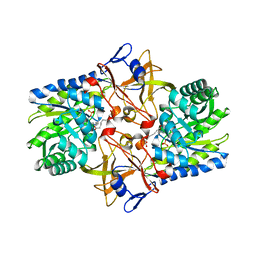 | | Crystal structure of Diaminopimelate Decarboxylase from m. jannaschi | | Descriptor: | AZELAIC ACID, Diaminopimelate decarboxylase | | Authors: | Rajashankar, K, Ray, S.R, Bonanno, J.B, Pinho, M.G, He, G, De Lencastre, H, Tomasz, A, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-06-24 | | Release date: | 2004-07-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Cocrystal structures of diaminopimelate decarboxylase: mechanism, evolution, and inhibition of an antibiotic resistance accessory factor
Structure, 10, 2002
|
|
1TUG
 
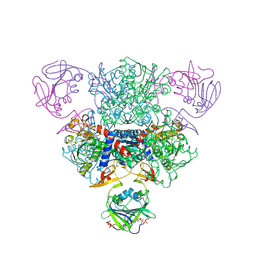 | | Aspartate Transcarbamoylase Catalytic Chain Mutant E50A Complex with Phosphonoacetamide, Malonate, and Cytidine-5-Prime-Triphosphate (CTP) | | Descriptor: | Aspartate carbamoyltransferase catalytic chain, Aspartate carbamoyltransferase regulatory chain, CYTIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Stieglitz, K, Stec, B, Baker, D.P, Kantrowitz, E.R. | | Deposit date: | 2004-06-24 | | Release date: | 2004-07-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Monitoring the Transition from the T to the R State in E.coli Aspartate Transcarbamoylase by X-ray Crystallography: Crystal Structures of the E50A Mutant Enzyme in Four Distinct Allosteric States.
J.Mol.Biol., 341, 2004
|
|
1TUH
 
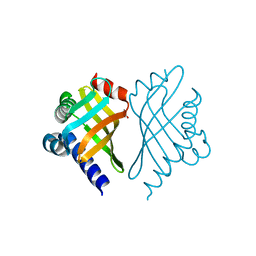 | | Structure of Bal32a from a Soil-Derived Mobile Gene Cassette | | Descriptor: | ACETATE ION, hypothetical protein EGC068 | | Authors: | Robinson, A, Wu, P.S.-C, Harrop, S.J, Schaeffer, P.M, Dixon, N.E, Gillings, M.R, Holmes, A.J, Nevalainen, K.M.H, Otting, G, Stokes, H.W, Curmi, P.M.G, Mabbutt, B.C. | | Deposit date: | 2004-06-25 | | Release date: | 2004-07-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Integron-associated Mobile Gene Cassettes Code for Folded Proteins: The Structure of Bal32a, a New Member of the Adaptable alpha+beta Barrel Family
J.Mol.Biol., 346, 2005
|
|
1TUI
 
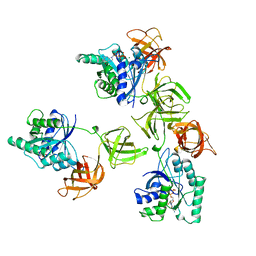 | | INTACT ELONGATION FACTOR TU IN COMPLEX WITH GDP | | Descriptor: | ELONGATION FACTOR TU, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Polekhina, G, Thirup, S, Kjeldgaard, M, Nissen, P, Lippmann, C, Nyborg, J. | | Deposit date: | 1996-05-23 | | Release date: | 1997-06-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Helix unwinding in the effector region of elongation factor EF-Tu-GDP.
Structure, 4, 1996
|
|
1TUJ
 
 | | Solution structure of the honey bee general odorant binding protein ASP2 in complex with trimethylsilyl-d4 propionate | | Descriptor: | 3-TRIMETHYLSILYL-PROPIONATE-2,2,3,3,-D4, odorant binding protein ASP2 | | Authors: | Lescop, E, Briand, L, Pernollet, J.-C, Guittet, E. | | Deposit date: | 2004-06-25 | | Release date: | 2005-09-20 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the honey bee general odorant binding protein ASP2 in complex with trimethylsilyl-d4 propionate
To be Published
|
|
1TUK
 
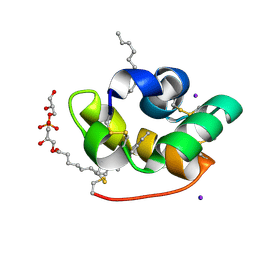 | | Crystal structure of liganded type 2 non specific lipid transfer protein from wheat | | Descriptor: | 1-MYRISTOYL-2-HYDROXY-SN-GLYCERO-3-[PHOSPHO-RAC-(1-GLYCEROL)], IODIDE ION, Nonspecific lipid-transfer protein 2G | | Authors: | Hoh, F, Pons, J.L, Gautier, M.F, De Lamotte, F, Dumas, C. | | Deposit date: | 2004-06-25 | | Release date: | 2005-04-05 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Structure of a liganded type 2 non-specific lipid-transfer protein from wheat and the molecular basis of lipid binding.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1TUL
 
 | | STRUCTURE OF TLP20 | | Descriptor: | TLP20 | | Authors: | Rayment, I, Holden, H.M. | | Deposit date: | 1996-08-17 | | Release date: | 1997-02-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular structure of a proteolytic fragment of TLP20.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
1TUM
 
 | | MUTT PYROPHOSPHOHYDROLASE-METAL-NUCLEOTIDE-METAL COMPLEX, NMR, 16 STRUCTURES | | Descriptor: | COBALT TETRAAMMINE ION, DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, MAGNESIUM ION, ... | | Authors: | Lin, J, Abeygunawardana, C, Frick, D.N, Bessman, M.J, Mildvan, A.S. | | Deposit date: | 1996-12-05 | | Release date: | 1997-05-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the quaternary MutT-M2+-AMPCPP-M2+ complex and mechanism of its pyrophosphohydrolase action.
Biochemistry, 36, 1997
|
|
1TUO
 
 | | Crystal structure of putative phosphomannomutase from Thermus Thermophilus HB8 | | Descriptor: | Putative phosphomannomutase | | Authors: | Misaki, S, Suzuki, S, Fujimoto, S, Sakurai, M, Kobayashi, M, Nishijima, K, Kunishima, N, Sugawara, M, Kuroishi, C, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-06-25 | | Release date: | 2005-08-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of putative phosphomannomutase from Thermus Thermophilus HB8
TO BE PUBLISHED
|
|
1TUP
 
 | | TUMOR SUPPRESSOR P53 COMPLEXED WITH DNA | | Descriptor: | DNA (5'-D(*AP*TP*AP*AP*TP*TP*GP*GP*GP*CP*AP*AP*GP*TP*CP*TP*A P*GP*GP*AP*A)-3'), DNA (5'-D(*TP*TP*TP*CP*CP*TP*AP*GP*AP*CP*TP*TP*GP*CP*CP*CP*A P*AP*TP*TP*A)-3'), PROTEIN (P53 TUMOR SUPPRESSOR ), ... | | Authors: | Cho, Y, Gorina, S, Jeffrey, P.D, Pavletich, N.P. | | Deposit date: | 1995-07-11 | | Release date: | 1995-07-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a p53 tumor suppressor-DNA complex: understanding tumorigenic mutations.
Science, 265, 1994
|
|
1TUQ
 
 | | NMR Structure Analysis of the B-DNA Dodecamer CTCtCACGTGGAG with a tricyclic cytosin base analogue | | Descriptor: | 5'-D(P*CP*TP*CP*(TC1)P*AP*CP*GP*TP*GP*GP*AP*G)-3' | | Authors: | Engman, K.C, Sandin, P, Osborne, S, Brown, T, Billeter, M, Lincoln, P, Norden, B, Albinsson, B, Wilhelmsson, L.M. | | Deposit date: | 2004-06-25 | | Release date: | 2004-10-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | DNA adopts normal B-form upon incorporation of highly fluorescent DNA base analogue tC: NMR structure and UV-Vis spectroscopy characterization.
Nucleic Acids Res., 32, 2004
|
|
1TUR
 
 | | SOLUTION STRUCTURE OF TURKEY OVOMUCOID THIRD DOMAIN AS DETERMINED FROM NUCLEAR MAGNETIC RESONANCE DATA | | Descriptor: | OVOMUCOID | | Authors: | Krezel, A.M, Darba, P, Robertson, A.D, Fejzo, J, Macura, S, Markley, J.L. | | Deposit date: | 1994-07-06 | | Release date: | 1994-10-15 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of turkey ovomucoid third domain as determined from nuclear magnetic resonance data.
J.Mol.Biol., 242, 1994
|
|
1TUS
 
 | |
1TUT
 
 | | J4/5 Loop from the Candida albicans and Candida dubliniensis Group I Introns | | Descriptor: | 5'-R(*GP*AP*GP*GP*AP*AP*GP*GP*CP*GP*A)-3', 5'-R(*UP*CP*GP*UP*UP*AP*AP*UP*CP*UP*C)-3' | | Authors: | Znosko, B.M, Kennedy, S.D, Wille, P.C, Krugh, T.R, Turner, D.H. | | Deposit date: | 2004-06-25 | | Release date: | 2004-12-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural Features and Thermodynamics of the J4/5 Loop from the Candida albicans and Candida dubliniensis Group I Introns.
Biochemistry, 43, 2004
|
|
1TUU
 
 | | Acetate Kinase crystallized with ATPgS | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, AMMONIUM ION, ... | | Authors: | Gorrell, A, Lawrence, S.H, Ferry, J.G. | | Deposit date: | 2004-06-25 | | Release date: | 2005-01-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and kinetic analyses of arginine residues in the active site of the acetate kinase from Methanosarcina thermophila.
J. Biol. Chem., 280, 2005
|
|
1TUV
 
 | | Crystal structure of YgiN in complex with menadione | | Descriptor: | MENADIONE, Protein ygiN | | Authors: | Adams, M.A, Jia, Z. | | Deposit date: | 2004-06-25 | | Release date: | 2005-01-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and Biochemical Evidence for an Enzymatic Quinone Redox Cycle in Escherichia coli: IDENTIFICATION OF A NOVEL QUINOL MONOOXYGENASE
J.Biol.Chem., 280, 2005
|
|
1TUW
 
 | | Structural and Functional Analysis of Tetracenomycin F2 Cyclase from Streptomyces glaucescens: A Type-II Polyketide Cyclase | | Descriptor: | SULFATE ION, Tetracenomycin polyketide synthesis protein tcmI | | Authors: | Thompson, T.B, Katayama, K, Watanabe, K, Hutchinson, C.R, Rayment, I. | | Deposit date: | 2004-06-25 | | Release date: | 2004-08-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and functional analysis of tetracenomycin F2 cyclase from Streptomyces glaucescens. A type II polyketide cyclase.
J.Biol.Chem., 279, 2004
|
|
1TUX
 
 | | HIGH RESOLUTION CRYSTAL STRUCTURE OF A THERMOSTABLE XYLANASE FROM THERMOASCUS AURANTIACUS | | Descriptor: | XYLANASE | | Authors: | Natesh, R, Bhanumoorthy, P, Vithayathil, P.J, Sekar, K, Ramakumar, S, Viswamitra, M.A. | | Deposit date: | 1998-10-29 | | Release date: | 1999-07-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure at 1.8 A resolution and proposed amino acid sequence of a thermostable xylanase from Thermoascus aurantiacus.
J.Mol.Biol., 288, 1999
|
|
1TUY
 
 | | Acetate Kinase complexed with ADP, AlF3 and acetate | | Descriptor: | ACETIC ACID, ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, ... | | Authors: | Gorrell, A, Lawrence, S.H, Ferry, J.G. | | Deposit date: | 2004-06-25 | | Release date: | 2005-01-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and Kinetic Analyses of Arginine Residues in the Active Site of the Acetate Kinase from Methanosarcina thermophila.
J.Biol.Chem., 280, 2005
|
|
1TUZ
 
 | | NMR Structure of the Diacylglycerol kinase alpha, NESGC target HR532 | | Descriptor: | diacylglycerol kinase alpha | | Authors: | Liu, G, Shao, Y, Xiao, R, Acton, T, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-06-25 | | Release date: | 2005-01-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of Diacylglycerol kinase alpha, NESGC target HR532
To be Published
|
|
1TV0
 
 | | Solution structure of cryptdin-4, the most potent alpha-defensin from mouse Paneth cells | | Descriptor: | Cryptdin-4 | | Authors: | Jing, W, Hunter, H.N, Tanabe, H, Ouellette, A.J, Vogel, H.J. | | Deposit date: | 2004-06-25 | | Release date: | 2005-01-04 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Cryptdin-4, a Mouse Paneth Cell alpha-Defensin.
Biochemistry, 43, 2004
|
|
