3NUD
 
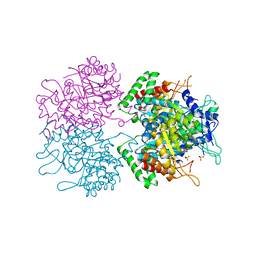 | | The structure of 3-deoxy-d-arabino-heptulosonate 7-phosphate synthase from mycobacterium tuberculosis complexed with phenylalanine | | Descriptor: | PHENYLALANINE, PHOSPHATE ION, Probable 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase AroG | | Authors: | Parker, E.J, Jameson, G.B, Jiao, W, Webby, C.J, Baker, E.N, Baker, H.M. | | Deposit date: | 2010-07-06 | | Release date: | 2010-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Synergistic allostery, a sophisticated regulatory network for the control of aromatic amino acid biosynthesis in Mycobacterium tuberculosis
J.Biol.Chem., 285, 2010
|
|
6EJB
 
 | |
4Q1L
 
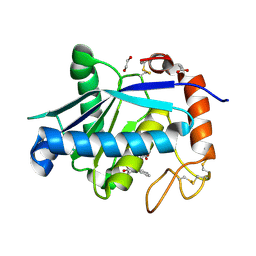 | | Crystal structure of Leucurolysin-a complexed with an endogenous tripeptide (QSW). | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Ferreira, R.N, Sanchez, E.O.F, Pimenta, A.M.C, Nagem, R.A.P. | | Deposit date: | 2014-04-03 | | Release date: | 2015-04-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of Leucurolysin-a complexed with an endogenous tripeptide (QSW).
To be Published
|
|
2SQC
 
 | |
5B0J
 
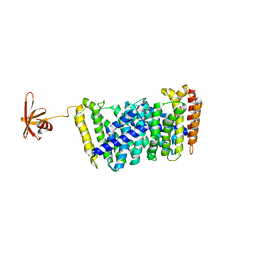 | | Structure of MoeN5-Sso7d fusion protein in complex with beta-undecyl maltoside | | Descriptor: | MoeN5,DNA-binding protein 7d, UNDECYL-MALTOSIDE | | Authors: | Ko, T.-P, Zhang, L, Chen, C.-C, Guo, R.-T, Oldfield, E.O. | | Deposit date: | 2015-10-30 | | Release date: | 2016-03-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Moenomycin Biosynthesis: Structure and Mechanism of Action of the Prenyltransferase MoeN5.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
2MHB
 
 | |
4HMM
 
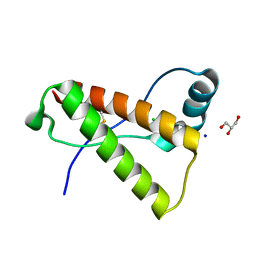 | | Crystal structure of mutant rabbit PRP 121-230 (S174N) | | Descriptor: | CHLORIDE ION, GLYCEROL, Major prion protein, ... | | Authors: | Sweeting, B, Chakrabartty, A, Pai, E.F. | | Deposit date: | 2012-10-18 | | Release date: | 2013-05-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | N-Terminal Helix-Cap in alpha-Helix 2 Modulates beta-State Misfolding in Rabbit and Hamster Prion Proteins.
Plos One, 8, 2013
|
|
3IK4
 
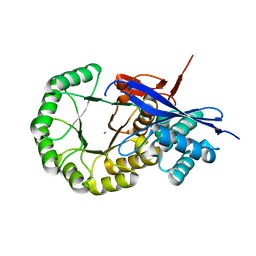 | | CRYSTAL STRUCTURE OF mandelate racemase/muconate lactonizing protein from Herpetosiphon aurantiacus | | Descriptor: | GLYCEROL, Mandelate racemase/muconate lactonizing protein, POTASSIUM ION | | Authors: | Patskovsky, Y, Toro, R, Dickey, M, Iizuka, M, Sauder, J.M, Gerlt, J.A, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-08-05 | | Release date: | 2009-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Homology models guide discovery of diverse enzyme specificities among dipeptide epimerases in the enolase superfamily.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
8OYD
 
 | |
6J8W
 
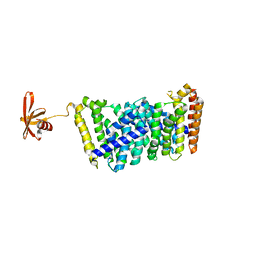 | | Structure of MOEN5-SSO7D fusion protein in complex with lig 1 | | Descriptor: | (2S)-3-dimethoxyphosphoryloxy-2-[(2E,6E)-3,7,11-trimethyldodeca-2,6,10-trienoxy]propanoic acid, MoeN5,DNA-binding protein 7d | | Authors: | Ko, T.P, Zhang, L.L, Chen, C.C, Guo, R.T. | | Deposit date: | 2019-01-21 | | Release date: | 2019-04-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Complex structures of MoeN5 with substrate analogues suggest sequential catalytic mechanism.
Biochem. Biophys. Res. Commun., 511, 2019
|
|
5ARH
 
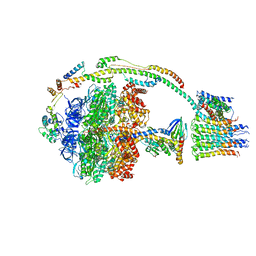 | | Bovine mitochondrial ATP synthase state 2a | | Descriptor: | ATP SYNTHASE F(0) COMPLEX SUBUNIT B1, MITOCHONDRIAL, ATP SYNTHASE F(0) COMPLEX SUBUNIT C1, ... | | Authors: | Zhou, A, Rohou, A, Schep, D.G, Bason, J.V, Montgomery, M.G, Walker, J.E, Grigorieff, N, Rubinstein, J.L. | | Deposit date: | 2015-09-24 | | Release date: | 2015-10-14 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.2 Å) | | Cite: | Structure and conformational states of the bovine mitochondrial ATP synthase by cryo-EM.
Elife, 4, 2015
|
|
4Q6J
 
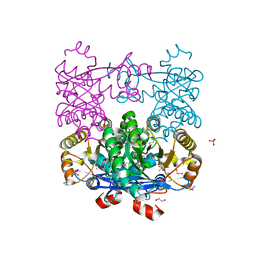 | | Crystal Structure of EAL domain Protein from Listeria monocytogenes EGD-e | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, GLYCEROL, ... | | Authors: | Kim, Y, Bigelow, L, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-04-22 | | Release date: | 2014-05-07 | | Method: | X-RAY DIFFRACTION (1.369 Å) | | Cite: | Crystal Structure of EAL domain Protein from Listeria monocytogenes EGD-e
To be Published
|
|
6EE8
 
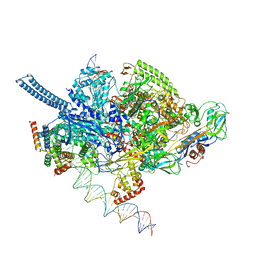 | | Mycobacterium tuberculosis RNAP promoter unwinding intermediate complex with RbpA/CarD and AP3 promoter | | Descriptor: | DNA (60-MER), DNA (65-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Darst, S.A, Campbell, E.A, Boyaci Selcuk, H, Chen, J. | | Deposit date: | 2018-08-13 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.92 Å) | | Cite: | Structures of an RNA polymerase promoter melting intermediate elucidate DNA unwinding.
Nature, 565, 2019
|
|
3IN1
 
 | |
2RNS
 
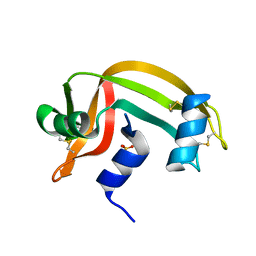 | | REFINEMENT OF THE CRYSTAL STRUCTURE OF RIBONUCLEASE S. COMPARISON WITH AND BETWEEN THE VARIOUS RIBONUCLEASE A STRUCTURES | | Descriptor: | RIBONUCLEASE S, SULFATE ION | | Authors: | Kim, E.E, Varadarajan, R, Wyckoff, H.W, Richards, F.M. | | Deposit date: | 1992-02-19 | | Release date: | 1994-01-31 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Refinement of the crystal structure of ribonuclease S. Comparison with and between the various ribonuclease A structures.
Biochemistry, 31, 1992
|
|
2HJH
 
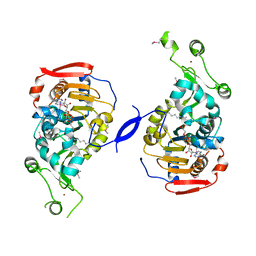 | | Crystal Structure of the Sir2 deacetylase | | Descriptor: | (2R,3R,4S,5R)-5-({[(R)-{[(R)-{[(2R,3S,4R,5R)-5-(6-AMINO-9H-PURIN-9-YL)-3,4-DIHYDROXYTETRAHYDROFURAN-2-YL]METHOXY}(HYDROXY)PHOSPHORYL]OXY}(HYDROXY)PHOSPHORYL]OXY}METHYL)-3,4-DIHYDROXYTETRAHYDROFURAN-2-YL ACETATE, NAD-dependent histone deacetylase SIR2, NICOTINAMIDE, ... | | Authors: | Hall, B.E, Ellenberger, T.E. | | Deposit date: | 2006-06-30 | | Release date: | 2008-04-08 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Autoregulation of the yeast Sir2 deacetylase by reaction and trapping of a pseudosubstrate motif in the active site
To be Published
|
|
6EJ7
 
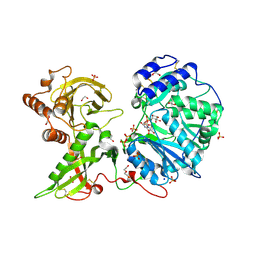 | |
1IM1
 
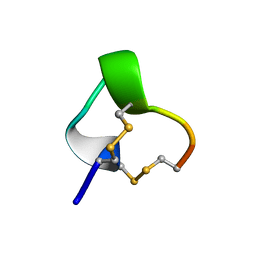 | | NMR SOLUTION STRUCTURE OF ALPHA-CONOTOXIN IM1, 20 STRUCTURES | | Descriptor: | ALPHA-CONOTOXIN IM1 | | Authors: | Rogers, J.P, Luginbuhl, P, Shen, G.S, Mccabe, R.T, Stevens, R.C, Wemmer, D.E. | | Deposit date: | 1998-11-18 | | Release date: | 1999-06-15 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of alpha-conotoxin ImI and comparison to other conotoxins specific for neuronal nicotinic acetylcholine receptors.
Biochemistry, 38, 1999
|
|
5ARA
 
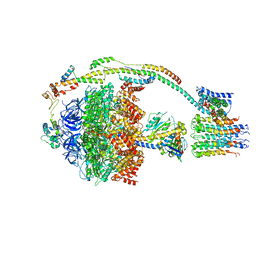 | | Bovine mitochondrial ATP synthase state 1a | | Descriptor: | ATP SYNTHASE F(0) COMPLEX SUBUNIT B1, MITOCHONDRIAL, ATP SYNTHASE F(0) COMPLEX SUBUNIT C1, ... | | Authors: | Zhou, A, Rohou, A, Schep, D.G, Bason, J.V, Montgomery, M.G, Walker, J.E, Grigorieff, N, Rubinstein, J.L. | | Deposit date: | 2015-09-24 | | Release date: | 2015-10-14 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.4 Å) | | Cite: | Structure and conformational states of the bovine mitochondrial ATP synthase by cryo-EM.
Elife, 4, 2015
|
|
3EQB
 
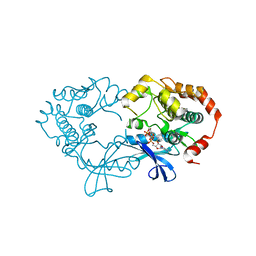 | | X-ray structure of the human mitogen-activated protein kinase kinase 1 (MEK1) in a complex with ligand and MgATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Dual specificity mitogen-activated protein kinase kinase 1, MAGNESIUM ION, ... | | Authors: | Ohren, J.F, Pavlovsky, A, Zhang, E. | | Deposit date: | 2008-09-30 | | Release date: | 2008-11-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | 2-Alkylamino- and alkoxy-substituted 2-amino-1,3,4-oxadiazoles-O-Alkyl benzohydroxamate esters replacements retain the desired inhibition and selectivity against MEK (MAP ERK kinase).
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3O5B
 
 | | Crystal structure of dimeric KlHxk1 in crystal form VII with glucose bound (open state) | | Descriptor: | Hexokinase, SULFATE ION, beta-D-glucopyranose | | Authors: | Kuettner, E.B, Kettner, K, Keim, A, Kriegel, T.M, Strater, N. | | Deposit date: | 2010-07-28 | | Release date: | 2010-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal Structure of Hexokinase KlHxk1 of Kluyveromyces lactis: A MOLECULAR BASIS FOR UNDERSTANDING THE CONTROL OF YEAST HEXOKINASE FUNCTIONS VIA COVALENT MODIFICATION AND OLIGOMERIZATION.
J.Biol.Chem., 285, 2010
|
|
3NR3
 
 | | Crystal Structure of Human Peripheral Myelin Protein 2 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, CITRIC ACID, ... | | Authors: | Ugochukwu, E, Pilka, E, Phillips, C, Yue, W.W, Krojer, T, von Delft, F, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A, Kavanagh, K.L, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-06-30 | | Release date: | 2010-08-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of Human Peripheral Myelin Protein 2
To be Published
|
|
1IYM
 
 | | RING-H2 finger domain of EL5 | | Descriptor: | EL5, ZINC ION | | Authors: | Katoh, E, Katoh, S, Minami, E, Yamazaki, T. | | Deposit date: | 2002-08-30 | | Release date: | 2003-07-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | High Precision NMR Structure and Function of the RING-H2 Finger Domain of EL5, a Rice Protein Whose Expression Is Increased upon Exposure to Pathogen-derived Oligosaccharides
J.Biol.Chem., 278, 2003
|
|
8ESG
 
 | | Bile Salt Hydrolase B from Lactobacillus gasseri with covalent inhibitor bound | | Descriptor: | (1R,3aS,3bR,5aR,7R,9aS,9bS,11aR)-1-[(2R)-6-fluoro-5-oxohexan-2-yl]-9a,11a-dimethylhexadecahydro-1H-cyclopenta[a]phenanthren-7-yl hydrogen sulfate (non-preferred name), Choloylglycine hydrolase | | Authors: | Walker, M.E, Redinbo, M.R. | | Deposit date: | 2022-10-14 | | Release date: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structural diversity of bile salt hydrolases reveals rationale for substrate selectivity
To Be Published
|
|
387D
 
 | | RNA Pseudoknot with 3D Domain Swapping | | Descriptor: | RNA Pseudoknot | | Authors: | Lietzke, S.E, Kundrot, C.E, Barnes, C.L. | | Deposit date: | 1998-04-14 | | Release date: | 2003-08-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The Structure of an RNA Pseudoknot Shows 3D Domain Swapping
Structure, Motion, Interaction and Expression of Biological Macromolecules, The Proceedings of the Tenth Conversation held at The University-SUNY, Albany NY, June 17-21, 1997, 10, 1998
|
|
