8QE4
 
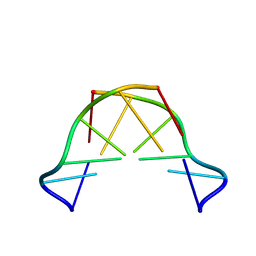 | | Formation of left-handed helices by C2'-fluorinated nucleic acids under physiological salt conditions | | Descriptor: | DNA (5'-D(*CP*(FRG)P*CP*(FRG)P*CP*(FRG))-3') | | Authors: | El-Khoury, R, Cabrero, C, Movilla, S, Friedland, D, Thorpe, J.D, Roman, M, Orozco, M, Gonzalez, C, Damha, M. | | Deposit date: | 2023-08-30 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-31 | | Method: | SOLUTION NMR | | Cite: | Formation of left-handed helices by C2'-fluorinated nucleic acids under physiological salt conditions.
Nucleic Acids Res., 52, 2024
|
|
8QE3
 
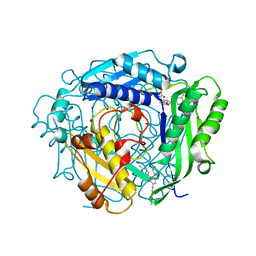 | | Crystal structure of human MAT2a bound to S-Adenosylmethionine and Compound 31 | | Descriptor: | 3-cyclopropyl-6-(2-methylindazol-5-yl)-4-(6-methylpyridin-3-yl)-2~{H}-pyrazolo[4,3-b]pyridin-5-one, S-ADENOSYLMETHIONINE, S-adenosylmethionine synthase isoform type-2 | | Authors: | Schimpl, M. | | Deposit date: | 2023-08-30 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.089 Å) | | Cite: | Development of a Series of Pyrrolopyridone MAT2A Inhibitors.
J.Med.Chem., 67, 2024
|
|
8QE2
 
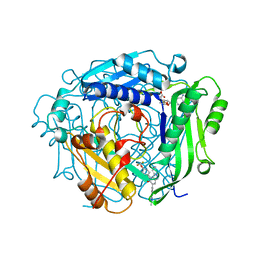 | | Crystal structure of human MAT2a bound to S-Adenosylmethionine and Compound 21 | | Descriptor: | 4-[4-[bis(fluoranyl)methoxy]phenyl]-3-cyclopropyl-6-(2-methylindazol-5-yl)-2~{H}-pyrazolo[4,3-b]pyridin-5-one, S-ADENOSYLMETHIONINE, S-adenosylmethionine synthase isoform type-2 | | Authors: | Schimpl, M. | | Deposit date: | 2023-08-30 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.109 Å) | | Cite: | Development of a Series of Pyrrolopyridone MAT2A Inhibitors.
J.Med.Chem., 67, 2024
|
|
8QE1
 
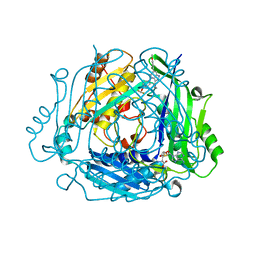 | | Crystal structure of human MAT2a bound to S-Adenosylmethionine and Compound 15 | | Descriptor: | 4-[4-[bis(fluoranyl)methoxy]phenyl]-3-cyclopropyl-6-(4-methoxyphenyl)-2~{H}-pyrazolo[4,3-b]pyridin-5-one, S-ADENOSYLMETHIONINE, S-adenosylmethionine synthase isoform type-2 | | Authors: | Schimpl, M. | | Deposit date: | 2023-08-30 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.095 Å) | | Cite: | Development of a Series of Pyrrolopyridone MAT2A Inhibitors.
J.Med.Chem., 67, 2024
|
|
8QE0
 
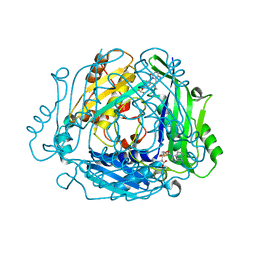 | | Crystal structure of human MAT2a bound to S-Adenosylmethionine and Compound 12 | | Descriptor: | 4-[4-[bis(fluoranyl)methoxy]phenyl]-3-cyclopropyl-2~{H}-pyrazolo[4,3-b]pyridin-5-one, S-ADENOSYLMETHIONINE, S-adenosylmethionine synthase isoform type-2 | | Authors: | Schimpl, M. | | Deposit date: | 2023-08-30 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Development of a Series of Pyrrolopyridone MAT2A Inhibitors.
J.Med.Chem., 67, 2024
|
|
8QDZ
 
 | |
8QDY
 
 | |
8QDU
 
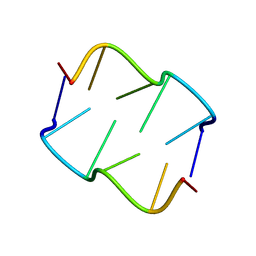 | | Formation of left-handed helices by C2'-fluorinated nucleic acids under physiological salt conditions | | Descriptor: | DNA (5'-D(*(FC)P*GP*(FC)P*GP*(FC)P*G)-3') | | Authors: | El-Khoury, R, Cabrero, C, Movilla, S, Thorpe, J.D, Roman, M, Orozco, M, Gonzalez, C, Damha, M.J. | | Deposit date: | 2023-08-30 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-31 | | Method: | SOLUTION NMR | | Cite: | Formation of left-handed helices by C2'-fluorinated nucleic acids under physiological salt conditions.
Nucleic Acids Res., 52, 2024
|
|
8QDH
 
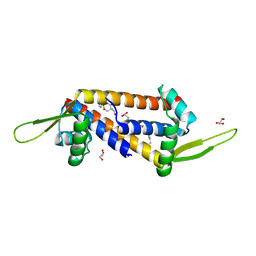 | | Engineered LmrR carrying a cyclic boronate ester formed between Tris and p-boronophenylalanine at position 89 | | Descriptor: | GLYCEROL, Transcriptional regulator, PadR-like family | | Authors: | Thunnissen, A.M.W.H, Rozeboom, H.J, Longwitz, L, Leveson-Gower, R.B, Roelfes, G. | | Deposit date: | 2023-08-29 | | Release date: | 2024-05-01 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Boron catalysis in a designer enzyme.
Nature, 629, 2024
|
|
8QDF
 
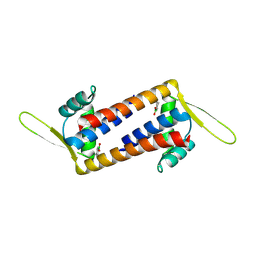 | | Engineered LmrR with Met-89 replaced by para-boronophenylalanine | | Descriptor: | Transcriptional regulator, PadR-like family | | Authors: | Thunnissen, A.M.W.H, Rozeboom, H.J, Longwitz, L, Leveson-Gower, R.B, Roelfes, G. | | Deposit date: | 2023-08-29 | | Release date: | 2024-05-01 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Boron catalysis in a designer enzyme.
Nature, 629, 2024
|
|
8QDD
 
 | | Structure of mBaoJin at pH 8.5 | | Descriptor: | CHLORIDE ION, SULFATE ION, mBaoJin | | Authors: | Samygina, V.R, Vlaskina, A.V, Gabdulkhakov, A, Subach, O.M, Subach, F.V. | | Deposit date: | 2023-08-28 | | Release date: | 2023-12-27 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Bright and stable monomeric green fluorescent protein derived from StayGold.
Nat.Methods, 21, 2024
|
|
8QDC
 
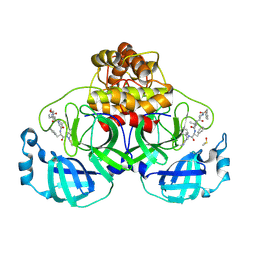 | | Crystal structure of SARS-CoV-2 main protease (MPro) in complex with the covalent inhibitor GUE-3642 (compound 1 in publication) | | Descriptor: | (phenylmethyl) ~{N}-[(2~{S})-1-[[(2~{S})-1-[[(2~{S})-1-[[iminomethyl-(phenylmethyl)amino]-methyl-amino]-1-oxidanylidene-3-phenyl-propan-2-yl]amino]-3,3-dimethyl-1-oxidanylidene-butan-2-yl]amino]-1-oxidanylidene-butan-2-yl]carbamate, 3C-like proteinase nsp5, CHLORIDE ION, ... | | Authors: | Strater, N, Claff, T, Sylvester, K, Mueller, C.E, Guetschow, M. | | Deposit date: | 2023-08-28 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Development of an active-site titrant for SARS-CoV-2 main protease as an indispensable tool for evaluating enzyme kinetics.
Acta Pharm Sin B, 14, 2024
|
|
8QCU
 
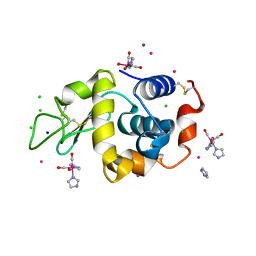 | | Lysozyme covalently bound to fac-[Re(CO)3-imidazole] complex, incubated for 38 weeks. | | Descriptor: | BROMIDE ION, CHLORIDE ION, IMIDAZOLE, ... | | Authors: | Jacobs, F.J.F, Brink, A, Helliwell, J.R. | | Deposit date: | 2023-08-28 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Time-series analysis of rhenium(I) organometallic covalent binding to a model protein for drug development.
Iucrj, 11, 2024
|
|
8QCK
 
 | |
8QCJ
 
 | |
8QCI
 
 | | FCGBP D10 Assembly Segment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GLYCEROL, ... | | Authors: | Yeshaya, N, Fass, D. | | Deposit date: | 2023-08-27 | | Release date: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | VWD domain stabilization by autocatalytic Asp-Pro cleavage.
Protein Sci., 33, 2024
|
|
8QC8
 
 | |
8QC4
 
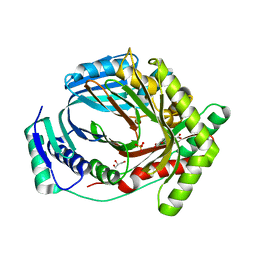 | | M. tuberculosis salicylate synthase MbtI in complex with 5-(3-carboxyphenyl)furan-2-carboxylic acid | | Descriptor: | 5-(3-carboxyphenyl)furan-2-carboxylic acid, GLYCEROL, SULFATE ION, ... | | Authors: | Mori, M, Villa, S, Meneghetti, F, Bellinzoni, M. | | Deposit date: | 2023-08-25 | | Release date: | 2023-11-15 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.578 Å) | | Cite: | Structural Study of a New MbtI-Inhibitor Complex: Towards an Optimized Model for Structure-Based Drug Discovery.
Pharmaceuticals, 16, 2023
|
|
8QC2
 
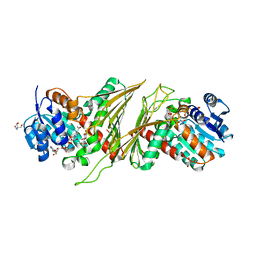 | | Crystal structure of NAD-dependent glycoside hydrolase from Flavobacterium sp. (strain K172) in complex with co-factor NAD+ and sulfoquinovose (SQ) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Gfo/Idh/MocA family oxidoreductase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Pickles, I.B, Sharma, M, Davies, G.J. | | Deposit date: | 2023-08-25 | | Release date: | 2023-12-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Widespread Family of NAD + -Dependent Sulfoquinovosidases at the Gateway to Sulfoquinovose Catabolism.
J.Am.Chem.Soc., 145, 2023
|
|
8QC0
 
 | |
8QBX
 
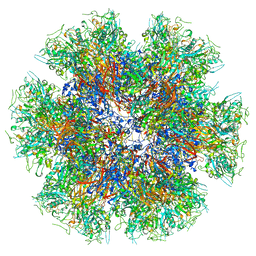 | | Chimeric Adenovirus-derived dodecamer | | Descriptor: | Penton protein | | Authors: | Buzas, D, Borucu, U, Bufton, J, Kapadalakere, S.Y, Toelzer, C. | | Deposit date: | 2023-08-25 | | Release date: | 2023-12-27 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Engineering the ADDobody protein scaffold for generation of high-avidity ADDomer super-binders.
Structure, 32, 2024
|
|
8QBP
 
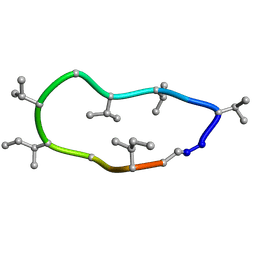 | | Conformations of macrocyclic peptides sampled by exact NOEs: models for cell-permeability. NMR structure of Omphalotin A in methanol / water indoleOut conformation. | | Descriptor: | TRP-MVA-ILE-MVA-MVA-SAR-MVA-IML-SAR-VAL-IML-SAR | | Authors: | Ruedisser, S.H, Matabaro, E, Sonderegger, L, Guentert, P, Kuenzler, M, Gossert, A.D. | | Deposit date: | 2023-08-25 | | Release date: | 2023-12-13 | | Last modified: | 2024-01-03 | | Method: | SOLUTION NMR | | Cite: | Conformations of Macrocyclic Peptides Sampled by Nuclear Magnetic Resonance: Models for Cell-Permeability.
J.Am.Chem.Soc., 145, 2023
|
|
8QBJ
 
 | | Structure of mBaoJin at pH 4.6 | | Descriptor: | CHLORIDE ION, mBaoJin | | Authors: | Samygina, V.R, Vlaskina, A.V, Gabdulkhakov, A, Subach, O.M, Subach, F.V. | | Deposit date: | 2023-08-24 | | Release date: | 2023-12-27 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Bright and stable monomeric green fluorescent protein derived from StayGold.
Nat.Methods, 21, 2024
|
|
8QB3
 
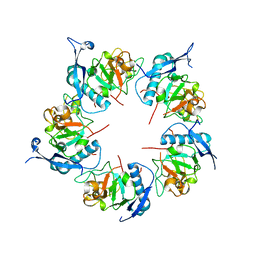 | | ADDobody zinc containing condition | | Descriptor: | ADDobody, ZINC ION | | Authors: | Buzas, D, Toelzer, C, Berger, I, Schaffitzel, C. | | Deposit date: | 2023-08-24 | | Release date: | 2023-12-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Engineering the ADDobody protein scaffold for generation of high-avidity ADDomer super-binders.
Structure, 32, 2024
|
|
8QB1
 
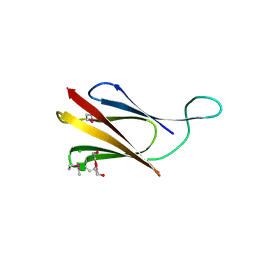 | | C-terminal domain of mirolase from Tannerella forsythia | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Mirolase, ... | | Authors: | Gomis-Ruth, F.X, Rodriguez-Banqueri, A, Mizgalska, D, Veillard, F, Goulas, T, Eckhard, U, Potempa, J. | | Deposit date: | 2023-08-23 | | Release date: | 2024-02-28 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Structural and functional insights into the C-terminal signal domain of the Bacteroidetes type-IX secretion system.
Open Biology, 14, 2024
|
|
