9DVK
 
 | | Structure of the phosphate exporter XPR1/SLC53A1, rotated dimer | | Descriptor: | 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CHOLESTEROL, PHOSPHATE ION, ... | | Authors: | Zhu, Q, Diver, M.M. | | Deposit date: | 2024-10-08 | | Release date: | 2025-04-02 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Transport and InsP 8 gating mechanisms of the human inorganic phosphate exporter XPR1.
Nat Commun, 16, 2025
|
|
9DVL
 
 | |
6RJF
 
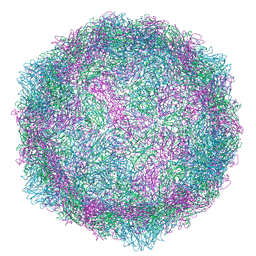 | | Echovirus 1 intact particle | | Descriptor: | PALMITIC ACID, VP1, VP2, ... | | Authors: | Domanska, A, Ruokolainen, V.P, Pelliccia, M, Laajala, M.A, Marjomaki, V.S, Butcher, S.J. | | Deposit date: | 2019-04-26 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Extracellular Albumin and Endosomal Ions Prime Enterovirus Particles for Uncoating That Can Be Prevented by Fatty Acid Saturation.
J.Virol., 93, 2019
|
|
6RKF
 
 | | Structure of human DASPO | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, D-aspartate oxidase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Chaves-Sanjuan, A, Nardini, M. | | Deposit date: | 2019-04-30 | | Release date: | 2020-03-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.219 Å) | | Cite: | Structure and kinetic properties of human d-aspartate oxidase, the enzyme-controlling d-aspartate levels in brain.
Faseb J., 34, 2020
|
|
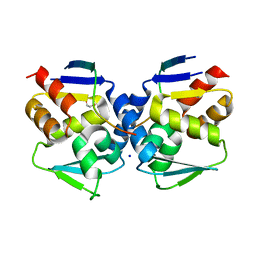
6XYX
 
 | |
6RTV
 
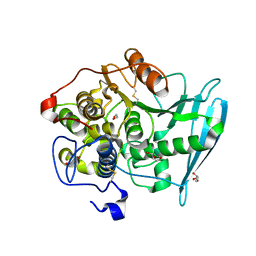 | | Crystal Structure of Glucuronoyl Esterase from Cerrena unicolor inactive S270A variant | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-O-methyl-glucuronoyl methylesterase, ... | | Authors: | Ernst, H.A, Mosbech, C, Langkilde, A, Westh, P, Meyer, A, Agger, J.W, Larsen, S. | | Deposit date: | 2019-05-27 | | Release date: | 2020-03-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | The structural basis of fungal glucuronoyl esterase activity on natural substrates.
Nat Commun, 11, 2020
|
|
8V1O
 
 | | Crystal structure of IRAK4 kinase domain with compound 4 | | Descriptor: | CHLORIDE ION, GLYCEROL, Interleukin-1 receptor-associated kinase 4, ... | | Authors: | Weiss, M.M, Zheng, X, Browne, C.M, Campbell, V, Chen, D, Enerson, B, Fei, X, Huang, X, Klaus, C.R, Li, H, Mayo, M, McDonald, A.A, Paul, A, Sharma, K, Shi, Y, Slavin, A, Walter, D.M, Yuan, K, Zhang, Y, Zhu, X, Kelleher, J, Ji, N, Walker, D, Mainolfi, N. | | Deposit date: | 2023-11-21 | | Release date: | 2024-07-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Discovery of KT-413, a Targeted Protein Degrader of IRAK4 and IMiD Substrates Targeting MYD88 Mutant Diffuse Large B-Cell Lymphoma.
J.Med.Chem., 67, 2024
|
|
5YJI
 
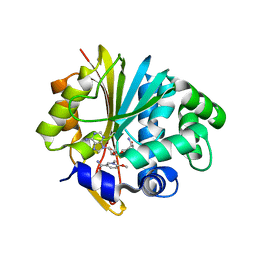 | | Co-crystal structure of Mouse Nicotinamide N-methyltransferase (NNMT) with small molecule analog of Nicotinamide | | Descriptor: | 6-methoxy-1-methyl-2H-pyridine-3-carboxamide, Nicotinamide N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Birudukota, S, Swaminathan, S, Thakur, M.K, Parveen, R, Kandan, S, Hallur, M.S, Rajagopal, S, Ruf, S, Dhakshinamoorthy, S, Kannt, A, Gosu, R. | | Deposit date: | 2017-10-10 | | Release date: | 2018-03-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | A small molecule inhibitor of Nicotinamide N-methyltransferase for the treatment of metabolic disorders.
Sci Rep, 8, 2018
|
|
7NY7
 
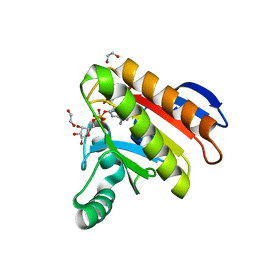 | | Crystal structure of the Capsaspora owczarzaki macroH2A macrodomain in complex with ADP-ribose | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5-DIPHOSPHORIBOSE, Histone macroH2A1.1 | | Authors: | Guberovic, I, Knobloch, G, Basquin, J, Buschbeck, M, Ladurner, A.G. | | Deposit date: | 2021-03-21 | | Release date: | 2021-11-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Evolution of a histone variant involved in compartmental regulation of NAD metabolism.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6IJ7
 
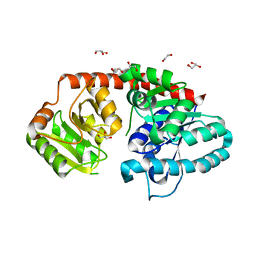 | | Crystal Structure of Arabidopsis thaliana UGT89C1 | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Rhamnosyltransferase protein | | Authors: | Zong, G, Wang, X. | | Deposit date: | 2018-10-09 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | Crystal structures of rhamnosyltransferase UGT89C1 from Arabidopsis thaliana reveal the molecular basis of sugar donor specificity for UDP-beta-l-rhamnose and rhamnosylation mechanism.
Plant J., 99, 2019
|
|
144D
 
 | | MINOR GROOVE BINDING OF SN6999 TO AN ALKYLATED DNA: MOLECULAR STRUCTURE OF D(CGC[E6G]AATTCGCG)-SN6999 COMPLEX | | Descriptor: | 1-METHYL-4-[4-[4-(4-(1-METHYLQUINOLINIUM)AMINO)BENZAMIDO]ANILINO]PYRIDINIUM, DNA (5'-D(*CP*GP*CP*(G36)P*AP*AP*TP*TP*CP*GP*CP*G)-3') | | Authors: | Gao, Y.-G, Sriram, M, Denny, W.A, Wang, A.H.-J. | | Deposit date: | 1993-10-26 | | Release date: | 1995-05-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Minor groove binding of SN6999 to an alkylated DNA: molecular structure of d(CGC[e6G]AATTCGCG)-SN6999 complex.
Biochemistry, 32, 1993
|
|
8SCW
 
 | |
1AAN
 
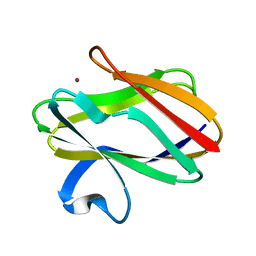 | | CRYSTAL STRUCTURE ANALYSIS OF AMICYANIN AND APOAMICYANIN FROM PARACOCCUS DENITRIFICANS AT 2.0 ANGSTROMS AND 1.8 ANGSTROMS RESOLUTION | | Descriptor: | AMICYANIN, COPPER (II) ION | | Authors: | Chen, L, Durley, R.C.E, Lim, L.W, Mathews, F.S. | | Deposit date: | 1992-04-09 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure analysis of amicyanin and apoamicyanin from Paracoccus denitrificans at 2.0 A and 1.8 A resolution.
Protein Sci., 2, 1993
|
|
5CLJ
 
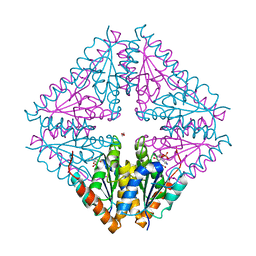 | |
8PFQ
 
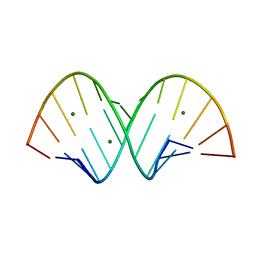 | | RNA structure with 1-methylpseudouridine, P1 space group | | Descriptor: | Chains: A,B, MAGNESIUM ION | | Authors: | Spingler, B, McAuley, K, Nievergelt, P, Thorn, A. | | Deposit date: | 2023-06-16 | | Release date: | 2024-01-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.01 Å) | | Cite: | RNA oligomers at atomic resolution containing 1-methylpseudouridine, an essential building block of mRNA vaccines.
Chemmedchem, 19, 2024
|
|
1AFK
 
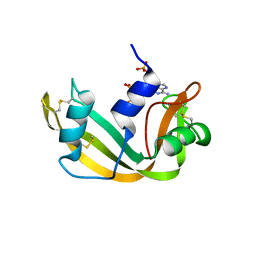 | |
5ZIQ
 
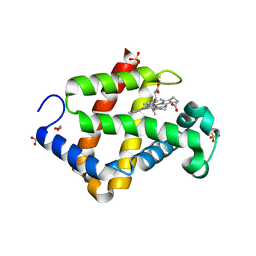 | | Crystal structure of hexacoordinated heme protein from anhydrobiotic tardigrade at pH 4 | | Descriptor: | 1,2-ETHANEDIOL, Globin protein, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Kim, J, Fukuda, Y, Inoue, T. | | Deposit date: | 2018-03-16 | | Release date: | 2019-01-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of Kumaglobin: a hexacoordinated heme protein from an anhydrobiotic tardigrade, Ramazzottius varieornatus.
FEBS J., 286, 2019
|
|
5O00
 
 | |
8PJ0
 
 | | Pseudomonas aeruginosa FabF C164A mutant in complex with N-(1,5-dimethyl-3-oxo-2-phenyl-2,3-dihydro-1H-pyrazol-4-yl)-3-methylbutanamide | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] synthase 2, DIMETHYL SULFOXIDE, FORMIC ACID, ... | | Authors: | Georgiou, C, Brenk, R, Espeland, L.O. | | Deposit date: | 2023-06-22 | | Release date: | 2024-02-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Design, quality and validation of the EU-OPENSCREEN fragment library poised to a high-throughput screening collection.
Rsc Med Chem, 15, 2024
|
|
9EOB
 
 | | Engineered GH181 sialidase from Akkermansia muciniphila | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Pichler, M.J, Banerjee, S, Nielsen, T.S, Morth, J.P, Abou Hachem, M. | | Deposit date: | 2024-03-14 | | Release date: | 2025-03-26 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Engineered GH181 sialidase from Akkermansia muciniphila
Acs Catalysis, 2025
|
|
7CL0
 
 | | Crystal structure of human SIRT6 | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, NAD-dependent protein deacetylase sirtuin-6, ... | | Authors: | Song, K, Zhang, J. | | Deposit date: | 2020-07-20 | | Release date: | 2021-02-24 | | Last modified: | 2025-09-17 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Reply to: Binding site for MDL-801 on SIRT6.
Nat.Chem.Biol., 17, 2021
|
|
7CNY
 
 | | Crystal structure of 8PE bound PSD from E. coli (2.12 A) | | Descriptor: | 1,2-Dioctanoyl-SN-Glycero-3-Phosphoethanolamine, DODECYL-BETA-D-MALTOSIDE, Phosphatidylserine decarboxylase alpha chain, ... | | Authors: | Kim, J, Cho, G. | | Deposit date: | 2020-08-03 | | Release date: | 2021-03-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structural insights into phosphatidylethanolamine formation in bacterial membrane biogenesis.
Sci Rep, 11, 2021
|
|
2VUQ
 
 | | Crystal structure of a human tRNAGly acceptor stem microhelix (derived from the gene sequence DG9990) at 1.18 Angstroem resolution | | Descriptor: | 5'-R(*CP*CP*AP*AP*UP*GP*CP)-3', 5'-R(*GP*CP*AP*UP*UP*GP*GP)-3' | | Authors: | Eichert, A, Perbandt, M, Schreiber, A, Fuerste, J.P, Betzel, C, Erdmann, V.A, Foerster, C. | | Deposit date: | 2008-05-29 | | Release date: | 2009-03-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Crystal Structure of the Human Trnagly Microhelix Isoacceptor G9990 at 1.18 A Resolution
Biochem.Biophys.Res.Commun., 380, 2009
|
|
5VM0
 
 | | The hapten triclocarban bound to the single domain camelid nanobody VHH T9 | | Descriptor: | 1,2-ETHANEDIOL, Camelid Nanobody VHH T9, N-(4-chlorophenyl)-N'-(3,4-dichlorophenyl)urea | | Authors: | Wogulis, L.A, Wogulis, M.D, Tabares-da Rosa, S, Gonzalez-Sapienza, G, Wilson, D.K. | | Deposit date: | 2017-04-26 | | Release date: | 2018-05-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structure and specificity of several triclocarban-binding single domain camelid antibody fragments.
J. Mol. Recognit., 32, 2019
|
|
5CXR
 
 | |
