9YFB
 
 | |
9YH9
 
 | |
4TZ5
 
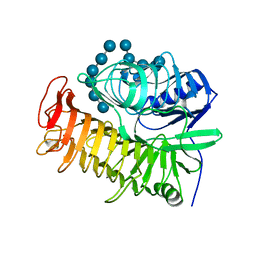 | | Ensemble refinement of the E502A variant of sacteLam55A from Streptomyces sp. SirexAA-E in complex with laminarihexaose | | Descriptor: | 1,2-ETHANEDIOL, Putative secreted protein, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose, ... | | Authors: | Bianchetti, C.M, Takasuka, T.E, Yik, E.J, Bergeman, L.F, Fox, B.G. | | Deposit date: | 2014-07-09 | | Release date: | 2015-03-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Active site and laminarin binding in glycoside hydrolase family 55.
J.Biol.Chem., 290, 2015
|
|
6J50
 
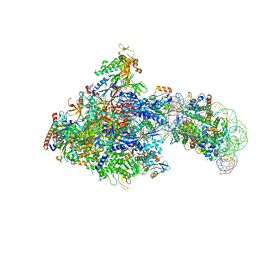 | | RNA polymerase II elongation complex bound with Spt4/5 and foreign DNA, stalled at SHL(-1) of the nucleosome (tilted conformation) | | Descriptor: | DNA (198-MER), DNA (41-MER), DNA-directed RNA polymerase subunit, ... | | Authors: | Ehara, H, Kujirai, T, Fujino, Y, Shirouzu, M, Kurumizaka, H, Sekine, S. | | Deposit date: | 2019-01-10 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Structural insight into nucleosome transcription by RNA polymerase II with elongation factors.
Science, 363, 2019
|
|
4UO0
 
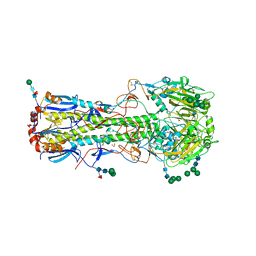 | | Structure of the A_Equine_Richmond_07 H3 haemagglutinin | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Vachieri, S.G, Collins, P.J, Haire, L.F, Ogrodowicz, R.W, Martin, S.R, Walker, P.A, Xiong, X, Gamblin, S.J, Skehel, J.J. | | Deposit date: | 2014-05-31 | | Release date: | 2014-07-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Recent Evolution of Equine Influenza and the Origin of Canine Influenza.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
6J4Z
 
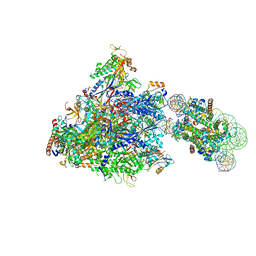 | | RNA polymerase II elongation complex bound with Spt4/5 and foreign DNA, stalled at SHL(-1) of the nucleosome | | Descriptor: | DNA (198-MER), DNA (42-MER), DNA-directed RNA polymerase subunit, ... | | Authors: | Ehara, H, Kujirai, T, Fujino, Y, Shirouzu, M, Kurumizaka, H, Sekine, S. | | Deposit date: | 2019-01-10 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural insight into nucleosome transcription by RNA polymerase II with elongation factors.
Science, 363, 2019
|
|
6J51
 
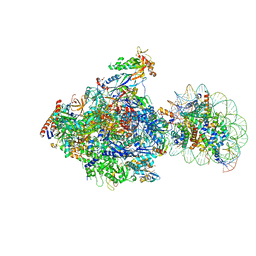 | | RNA polymerase II elongation complex bound with Spt4/5 and foreign DNA, stalled at SHL(-1) of the nucleosome, weak Elf1 (+1 position) | | Descriptor: | DNA (198-MER), DNA (36-MER), DNA-directed RNA polymerase subunit, ... | | Authors: | Ehara, H, Kujirai, T, Fujino, Y, Shirouzu, M, Kurumizaka, H, Sekine, S. | | Deposit date: | 2019-01-10 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural insight into nucleosome transcription by RNA polymerase II with elongation factors.
Science, 363, 2019
|
|
6JP8
 
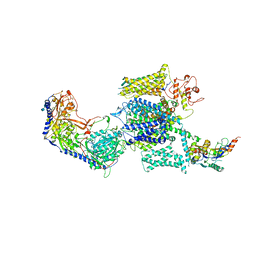 | | Rabbit Cav1.1-Bay K8644 Complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhao, Y, Huang, G, Wu, J, Yan, N. | | Deposit date: | 2019-03-26 | | Release date: | 2019-06-12 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Molecular Basis for Ligand Modulation of a Mammalian Voltage-Gated Ca2+Channel.
Cell, 177, 2019
|
|
8XC7
 
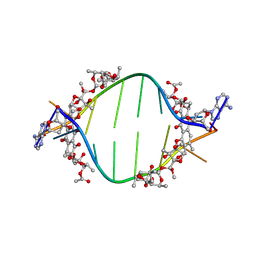 | |
9YFK
 
 | |
9V14
 
 | | crystal structure of the enzyme-product complex of L-azetidine-2-carboxylate hydrolase | | Descriptor: | 1,2-ETHANEDIOL, 4-AMINO-2-HYDROXYBUTANOIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Toyoda, M, Mizutani, K, Mikami, B, Wackett, L.P, Esaki, N, Kurihara, T. | | Deposit date: | 2025-05-19 | | Release date: | 2025-06-04 | | Method: | X-RAY DIFFRACTION (0.93 Å) | | Cite: | Research for the crystal structure of L-azetidine-2-carboxylate hydrolase
To Be Published
|
|
9O0X
 
 | | Co-crystal structure of human TREX1 in complex with an inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 5-({N-[(2,3-dichlorophenyl)methyl]-3,5-difluorobenzamido}methyl)-3-(methylamino)pyridine-2-carboxylic acid, MAGNESIUM ION, ... | | Authors: | Dehghani-Tafti, S, Dong, A, Li, Y, Ackloo, S, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2025-04-03 | | Release date: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Co-crystal structure of human TREX1 in complex with an inhibitor
To be published
|
|
9O0W
 
 | | Co-crystal structure of human TREX1 in complex with an inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 5-({N-[(2,3-dichlorophenyl)methyl]-3,5-difluorobenzamido}methyl)pyridine-2-carboxylic acid, MAGNESIUM ION, ... | | Authors: | Dehghani-Tafti, S, Dong, A, Li, Y, Ackloo, S, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2025-04-03 | | Release date: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Co-crystal structure of human TREX1 in complex with an inhibitor
To be published
|
|
9VGE
 
 | | SIRT2 demyristoylation intermediate I structure | | Descriptor: | GLYCEROL, Histone H3.1, NAD-dependent protein deacetylase sirtuin-2, ... | | Authors: | Zhang, N, Hao, Q. | | Deposit date: | 2025-06-13 | | Release date: | 2025-09-24 | | Last modified: | 2025-10-01 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Structural basis of SIRT2 pre-catalysis NAD + binding dynamics and mechanism.
Rsc Chem Biol, 2025
|
|
6W5G
 
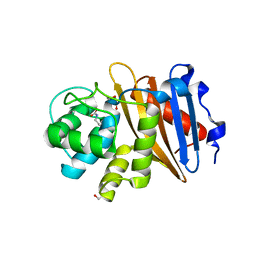 | | Class D beta-lactamase BAT-2 | | Descriptor: | 1,2-ETHANEDIOL, BAT-2 beta-lactamase | | Authors: | Smith, C.A, Vakulenko, S.B, Stewart, N.K, Toth, M. | | Deposit date: | 2020-03-13 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.451 Å) | | Cite: | A surface loop modulates activity of the Bacillus class D beta-lactamases.
J.Struct.Biol., 211, 2020
|
|
9Y81
 
 | |
9UI2
 
 | |
9E61
 
 | | Cryo-EM structure of Saccharomyces cerevisiae Pmt4 apo form | | Descriptor: | 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Dolichyl-phosphate-mannose--protein mannosyltransferase 4 | | Authors: | Du, M, Yuan, Z, Li, H. | | Deposit date: | 2024-10-29 | | Release date: | 2025-10-15 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Pmt4 recognizes two separate acceptor sites to O-mannosylate the S/T-rich regions of substrate proteins
To Be Published
|
|
9E6V
 
 | | Cryo-EM structure of Saccharomyces cerevisiae Pmt4-Ccw5 complex | | Descriptor: | 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Cell wall mannoprotein CIS3, Dolichyl-phosphate-mannose--protein mannosyltransferase 4, ... | | Authors: | Du, M, Yuan, Z, Li, H. | | Deposit date: | 2024-10-31 | | Release date: | 2025-10-15 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Pmt4 recognizes two separate acceptor sites to O-mannosylate the S/T-rich regions of substrate proteins
To Be Published
|
|
115D
 
 | | ORDERED WATER STRUCTURE IN AN A-DNA OCTAMER AT 1.7 ANGSTROMS RESOLUTION | | Descriptor: | DNA (5'-D(*GP*GP*(BRU)P*AP*(BRU)P*AP*CP*C)-3') | | Authors: | Kennard, O, Cruse, W.B.T, Nachman, J, Prange, T, Shakked, Z, Rabinovich, D. | | Deposit date: | 1993-02-12 | | Release date: | 1993-07-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Ordered water structure in an A-DNA octamer at 1.7 A resolution.
J.Biomol.Struct.Dyn., 3, 1986
|
|
9VCK
 
 | | Cryo-EM structure of SARS-CoV-2 nsp10/nsp14:RNA:SMP complex | | Descriptor: | CALCIUM ION, Guanine-N7 methyltransferase nsp14, Non-structural protein 10, ... | | Authors: | Wang, J, Lou, Z, Liu, D. | | Deposit date: | 2025-06-06 | | Release date: | 2025-07-16 | | Method: | ELECTRON MICROSCOPY (4.22 Å) | | Cite: | Structural Basis and Rational Design of Nucleotide Analogue Inhibitor Evading the SARS-CoV-2 Proofreading Enzyme.
J.Am.Chem.Soc., 147, 2025
|
|
9VCL
 
 | | Cryo-EM structure of SARS-CoV-2 nsp10/nsp14:RNA:ATMP complex | | Descriptor: | CALCIUM ION, Guanine-N7 methyltransferase nsp14, Non-structural protein 10, ... | | Authors: | Wang, J, Lou, Z, Liu, D. | | Deposit date: | 2025-06-06 | | Release date: | 2025-07-16 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Structural Basis and Rational Design of Nucleotide Analogue Inhibitor Evading the SARS-CoV-2 Proofreading Enzyme.
J.Am.Chem.Soc., 147, 2025
|
|
9QYT
 
 | | Crystal structure of leaf branch compost cutinase variant ICCG L50Y T110E | | Descriptor: | 1,2-ETHANEDIOL, Leaf-branch compost cutinase, SODIUM ION | | Authors: | Bischoff, D, Walla, B, Janowski, R, Niessing, D, Weuster-Botz, D. | | Deposit date: | 2025-04-20 | | Release date: | 2025-07-02 | | Last modified: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Application of a Rational Crystal Contact Engineering Strategy on a Poly(ethylene terephthalate)-Degrading Cutinase.
Bioengineering (Basel), 12, 2025
|
|
9RYG
 
 | |
9QYU
 
 | | Crystal structure of leaf branch compost cutinase quintuple variant ICCG L50Y | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Leaf-branch compost cutinase | | Authors: | Bischoff, D, Walla, B, Dietrich, A.-M, Janowski, R, Niessing, D, Weuster-Botz, D. | | Deposit date: | 2025-04-21 | | Release date: | 2025-07-02 | | Last modified: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Application of a Rational Crystal Contact Engineering Strategy on a Poly(ethylene terephthalate)-Degrading Cutinase.
Bioengineering (Basel), 12, 2025
|
|
