5ZEY
 
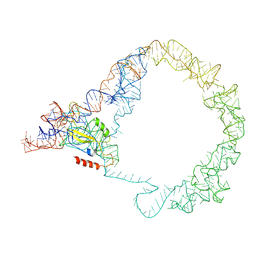 | | M. smegmatis Trans-translation state 70S ribosome | | Descriptor: | A-tRNAfMet, SsrA-binding protein, tmRNA | | Authors: | Mishra, S, Ahmed, T, Tyagi, A, Shi, J, Bhushan, S. | | Deposit date: | 2018-02-28 | | Release date: | 2018-09-26 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (12.5 Å) | | Cite: | Structures of Mycobacterium smegmatis 70S ribosomes in complex with HPF, tmRNA, and P-tRNA.
Sci Rep, 8, 2018
|
|
4X4R
 
 | |
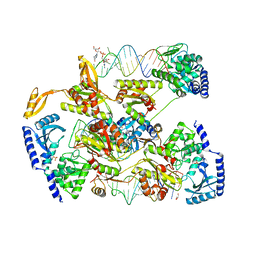
4X4N
 
 | |
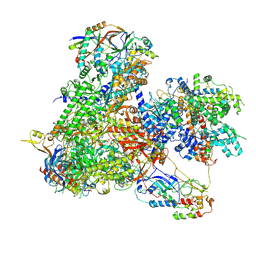
7Z30
 
 | |
7Z0H
 
 | | Structure of yeast RNA Polymerase III-Ty1 integrase complex at 2.6 A (focus subunit AC40). | | Descriptor: | DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, DNA-directed RNA polymerase III subunit RPC2, ... | | Authors: | Nguyen, P.Q, Huecas, S, Plaza-Pegueroles, A, Fernandez-Tornero, C. | | Deposit date: | 2022-02-22 | | Release date: | 2023-04-05 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural basis of Ty1 integrase tethering to RNA polymerase III for targeted retrotransposon integration.
Nat Commun, 14, 2023
|
|
7Z31
 
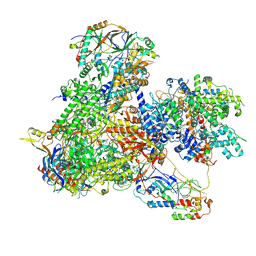 | | Structure of yeast RNA Polymerase III-Ty1 integrase complex at 2.7 A (focus subunit C11, no C11 C-terminal Zn-ribbon in the funnel pore). | | Descriptor: | DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, DNA-directed RNA polymerase III subunit RPC2, ... | | Authors: | Nguyen, P.Q, Huecas, S, Plaza-Pegueroles, A, Fernandez-Tornero, C. | | Deposit date: | 2022-03-01 | | Release date: | 2023-04-05 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Structural basis of Ty1 integrase tethering to RNA polymerase III for targeted retrotransposon integration.
Nat Commun, 14, 2023
|
|
4X4S
 
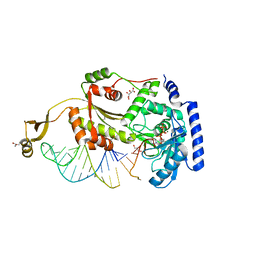 | |
4X4O
 
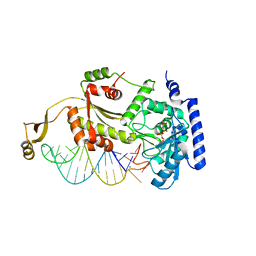 | |
8FNC
 
 | | Cryo-EM structure of RNase-treated RESC-C in trypanosomal RNA editing | | Descriptor: | Mitochondrial RNA binding complex 1 subunit, Mitochondrial RNA binding protein, Phytanoyl-CoA dioxygenase family protein, ... | | Authors: | Liu, S, Wang, H, Li, X, Zhang, F, Lee, J.K.J, Li, Z, Yu, C, Zhao, X, Hu, J.J, Suematsu, T, Alvarez-Cabrera, A.L, Liu, Q, Zhang, L, Huang, L, Aphasizheva, I, Aphasizhev, R, Zhou, Z.H. | | Deposit date: | 2022-12-27 | | Release date: | 2023-07-19 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of gRNA stabilization and mRNA recognition in trypanosomal RNA editing.
Science, 381, 2023
|
|
8FNF
 
 | | Cryo-EM structure of RNase-untreated RESC-C in trypanosomal RNA editing | | Descriptor: | Mitochondrial RNA binding complex 1 subunit, Mitochondrial RNA binding protein, Phytanoyl-CoA dioxygenase family protein, ... | | Authors: | Liu, S, Wang, H, Li, X, Zhang, F, Lee, J.K.J, Li, Z, Yu, C, Zhao, X, Hu, J.J, Suematsu, T, Alvarez-Cabrera, A.L, Liu, Q, Zhang, L, Huang, L, Aphasizheva, I, Aphasizhev, R, Zhou, Z.H. | | Deposit date: | 2022-12-27 | | Release date: | 2023-07-19 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of gRNA stabilization and mRNA recognition in trypanosomal RNA editing.
Science, 381, 2023
|
|
4X4P
 
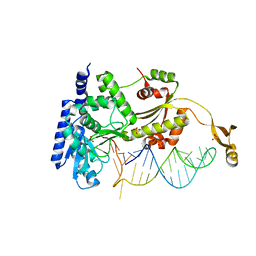 | |
4X4Q
 
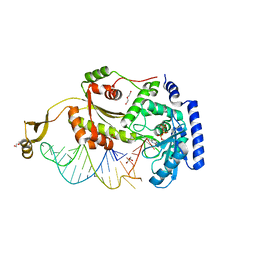 | |
8PV1
 
 | | Chaetomium thermophilum pre-60S State 6 - pre-5S rotation - L1 intermediate - composite structure | | Descriptor: | 26S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Thoms, M, Cheng, J, Denk, T, Berninghausen, O, Beckmann, R. | | Deposit date: | 2023-07-17 | | Release date: | 2023-11-15 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | Structural insights into coordinating 5S RNP rotation with ITS2 pre-RNA processing during ribosome formation.
Embo Rep., 24, 2023
|
|
8PV3
 
 | | Chaetomium thermophilum pre-60S State 9 - pre-5S rotation - immature H68/H69 - composite structure | | Descriptor: | 26S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Thoms, M, Cheng, J, Denk, T, Berninghausen, O, Beckmann, R. | | Deposit date: | 2023-07-17 | | Release date: | 2023-11-15 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural insights into coordinating 5S RNP rotation with ITS2 pre-RNA processing during ribosome formation.
Embo Rep., 24, 2023
|
|
8PV7
 
 | | Chaetomium thermophilum pre-60S State 1 - pre-5S rotation (Arx1/Nog2 state) - Composite structure | | Descriptor: | 26S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Thoms, M, Cheng, J, Denk, T, Berninghausen, O, Beckmann, R. | | Deposit date: | 2023-07-17 | | Release date: | 2023-11-15 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (2.12 Å) | | Cite: | Structural insights into coordinating 5S RNP rotation with ITS2 pre-RNA processing during ribosome formation.
Embo Rep., 24, 2023
|
|
8PVK
 
 | | Chaetomium thermophilum pre-60S State 5 - pre-5S rotation - L1 inward - composite structure | | Descriptor: | 26S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Thoms, M, Cheng, J, Denk, T, Berninghausen, O, Beckmann, R. | | Deposit date: | 2023-07-17 | | Release date: | 2023-12-06 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (2.55 Å) | | Cite: | Structural insights into coordinating 5S RNP rotation with ITS2 pre-RNA processing during ribosome formation.
Embo Rep., 24, 2023
|
|
8PVL
 
 | | Chaetomium thermophilum pre-60S State 7 - pre-5S rotation lacking Utp30/ITS2 - composite structure | | Descriptor: | 26S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Thoms, M, Cheng, J, Denk, T, Berninghausen, O, Beckmann, R. | | Deposit date: | 2023-07-17 | | Release date: | 2023-12-06 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (2.19 Å) | | Cite: | Structural insights into coordinating 5S RNP rotation with ITS2 pre-RNA processing during ribosome formation.
Embo Rep., 24, 2023
|
|
5CD1
 
 | | Structure of an asymmetric tetramer of human tRNA m1A58 methyltransferase in a complex with SAH and tRNA3Lys | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, SODIUM ION, tRNA (adenine(58)-N(1))-methyltransferase catalytic subunit TRMT61A, ... | | Authors: | Finer-Moore, J, Czudnochowski, N, O'Connell III, J.D, Wang, A.L, Stroud, R.M. | | Deposit date: | 2015-07-02 | | Release date: | 2015-11-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Crystal Structure of the Human tRNA m(1)A58 Methyltransferase-tRNA3(Lys) Complex: Refolding of Substrate tRNA Allows Access to the Methylation Target.
J.Mol.Biol., 427, 2015
|
|
5CCX
 
 | | Structure of the product complex of tRNA m1A58 methyltransferase with tRNA3Lys as substrate | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, SODIUM ION, tRNA (adenine(58)-N(1))-methyltransferase catalytic subunit TRMT61A, ... | | Authors: | Finer-Moore, J, Czudnochowski, N, O'Connell III, J.D, Wang, A.L, Stroud, R.M. | | Deposit date: | 2015-07-02 | | Release date: | 2015-11-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of the Human tRNA m(1)A58 Methyltransferase-tRNA3(Lys) Complex: Refolding of Substrate tRNA Allows Access to the Methylation Target.
J.Mol.Biol., 427, 2015
|
|
5CCB
 
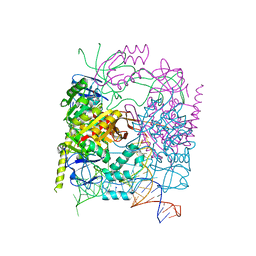 | | Crystal structure of human m1A58 methyltransferase in a complex with tRNA3Lys and SAH | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, SODIUM ION, tRNA (adenine(58)-N(1))-methyltransferase catalytic subunit TRMT61A, ... | | Authors: | Finer-Moore, J, Czudnochowski, N, O'Connell III, J.D, Wang, A.L, Stroud, R.M. | | Deposit date: | 2015-07-01 | | Release date: | 2015-11-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Human tRNA m(1)A58 Methyltransferase-tRNA3(Lys) Complex: Refolding of Substrate tRNA Allows Access to the Methylation Target.
J.Mol.Biol., 427, 2015
|
|
8PV2
 
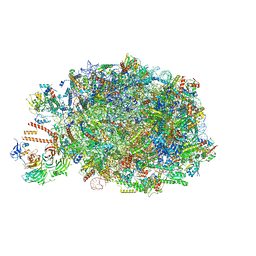 | | Chaetomium thermophilum pre-60S State 10 - pre-5S rotation with Ytm1-Erb1 | | Descriptor: | 26S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Thoms, M, Cheng, J, Denk, T, Berninghausen, O, Beckmann, R. | | Deposit date: | 2023-07-17 | | Release date: | 2023-11-15 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (2.63 Å) | | Cite: | Structural insights into coordinating 5S RNP rotation with ITS2 pre-RNA processing during ribosome formation.
Embo Rep., 24, 2023
|
|
6OF4
 
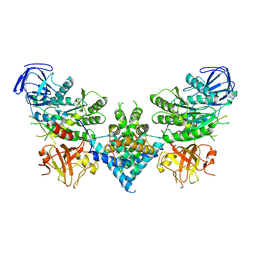 | | Precursor ribosomal RNA processing complex, apo-state. | | Descriptor: | CLP1_P domain-containing protein, Ribonuclease | | Authors: | Pillon, M.C, Hsu, A.L, Krahn, J.M, Williams, J.G, Goslen, K.H, Sobhany, M, Borgnia, M.J, Stanley, R.E. | | Deposit date: | 2019-03-28 | | Release date: | 2019-09-11 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM reveals active site coordination within a multienzyme pre-rRNA processing complex.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6OF2
 
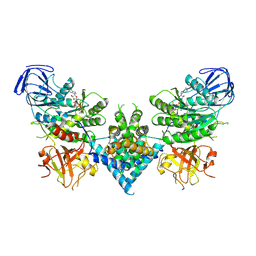 | | Precursor ribosomal RNA processing complex, State 2. | | Descriptor: | CLP1_P domain-containing protein, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Pillon, M.C, Hsu, A.L, Krahn, J.M, Williams, J.G, Goslen, K.H, Sobhany, M, Borgnia, M.J, Stanley, R.E. | | Deposit date: | 2019-03-28 | | Release date: | 2019-09-11 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM reveals active site coordination within a multienzyme pre-rRNA processing complex.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6OF3
 
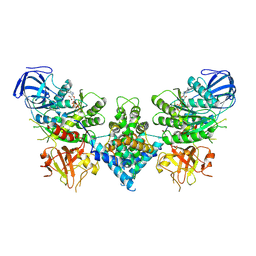 | | Precursor ribosomal RNA processing complex, State 1. | | Descriptor: | CLP1_P domain-containing protein, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Pillon, M.C, Hsu, A.L, Krahn, J.M, Williams, J.G, Goslen, K.H, Sobhany, M, Borgnia, M.J, Stanley, R.E. | | Deposit date: | 2019-03-28 | | Release date: | 2019-09-11 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM reveals active site coordination within a multienzyme pre-rRNA processing complex.
Nat.Struct.Mol.Biol., 26, 2019
|
|
8PV5
 
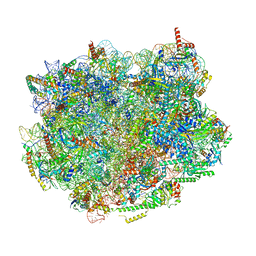 | | Chaetomium thermophilum pre-60S State 8 - pre-5S rotation without Foot - composite structure | | Descriptor: | 26S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Thoms, M, Cheng, J, Denk, T, Berninghausen, O, Beckmann, R. | | Deposit date: | 2023-07-17 | | Release date: | 2023-11-15 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Structural insights into coordinating 5S RNP rotation with ITS2 pre-RNA processing during ribosome formation.
Embo Rep., 24, 2023
|
|
