7GZX
 
 | | Crystal structure of SARS-CoV-2 NSP3 Macrodomain in complex with ASAP-0011176-001 | | Descriptor: | 7-[(1S)-2-methyl-1-{[(6M)-6-{5-[(methylamino)methyl]furan-3-yl}-7H-pyrrolo[2,3-d]pyrimidin-4-yl]amino}propyl]-3,4-dihydro-1lambda~6~-benzothiopyran-1,1(2H)-dione, Papain-like protease nsp3 | | Authors: | Aschenbrenner, J.C, Fearon, D, Tomlinson, C.W.E, Marples, P.G, Fairhead, M, Balcomb, B.H, Chandran, A.V, Godoy, A.S, Koekemoer, L, Lithgo, R.M, Ni, X, Thompson, W, Wang, S, Wild, C, Williams, E.P, Winokan, M, Walsh, M.A, von Delft, F. | | Deposit date: | 2024-01-23 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Group deposition of SARS-CoV-2 NSP3 Macrodomain in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
7H14
 
 | | Crystal structure of SARS-CoV-2 NSP3 Macrodomain in complex with ASAP-0013383-001 | | Descriptor: | 7-[(1R)-2-methyl-1-{[(6M)-6-(1-methyl-6-oxo-1,6-dihydropyridin-3-yl)-7H-pyrrolo[2,3-d]pyrimidin-4-yl]amino}propyl]-3,4-dihydro-1lambda~6~-benzothiopyran-1,1(2H)-dione, Papain-like protease nsp3 | | Authors: | Aschenbrenner, J.C, Fearon, D, Tomlinson, C.W.E, Marples, P.G, Fairhead, M, Balcomb, B.H, Chandran, A.V, Godoy, A.S, Koekemoer, L, Lithgo, R.M, Ni, X, Thompson, W, Wang, S, Wild, C, Williams, E.P, Winokan, M, Walsh, M.A, von Delft, F. | | Deposit date: | 2024-01-23 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Group deposition of SARS-CoV-2 NSP3 Macrodomain in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
7H1F
 
 | | Crystal structure of SARS-CoV-2 NSP3 Macrodomain in complex with ASAP-0015381-001 | | Descriptor: | N-[(1R)-1-(4-methoxyphenyl)-2-(1H-tetrazol-5-yl)ethyl]-7H-pyrrolo[2,3-d]pyrimidine-4-carboxamide, Papain-like protease nsp3 | | Authors: | Aschenbrenner, J.C, Fearon, D, Tomlinson, C.W.E, Marples, P.G, Fairhead, M, Balcomb, B.H, Chandran, A.V, Godoy, A.S, Koekemoer, L, Lithgo, R.M, Ni, X, Thompson, W, Wang, S, Wild, C, Williams, E.P, Winokan, M, Walsh, M.A, von Delft, F. | | Deposit date: | 2024-01-23 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Group deposition of SARS-CoV-2 NSP3 Macrodomain in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
4GFT
 
 | | Malaria invasion machinery protein-Nanobody complex | | Descriptor: | 1,2-ETHANEDIOL, Myosin A tail domain interacting protein, Nanobody | | Authors: | Khamrui, S, Turley, S, Pardon, E, Steyaert, J, Verlinde, C, Fan, E, Bergman, L.W, Hol, W.G.J. | | Deposit date: | 2012-08-03 | | Release date: | 2013-07-03 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The structure of the D3 domain of Plasmodium falciparum myosin tail interacting protein MTIP in complex with a nanobody.
Mol.Biochem.Parasitol., 190, 2013
|
|
5KCF
 
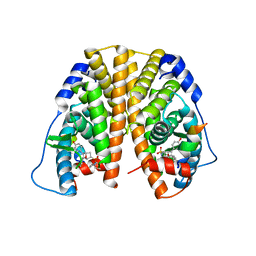 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with an N-ethyl, 4-methoxybenzyl OBHS-N derivative | | Descriptor: | (1R,2S,4R)-N-ethyl-5,6-bis(4-hydroxyphenyl)-N-(4-methoxyphenyl)-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonamide, (1S,2R,4S)-N-ethyl-5,6-bis(4-hydroxyphenyl)-N-(4-methoxyphenyl)-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonamide, Estrogen receptor, ... | | Authors: | Nwachukwu, J.C, Srinivasan, S, Bruno, N.E, Dharmarajan, V, Goswami, D, Kastrati, I, Novick, S, Nowak, J, Zhou, H.B, Boonmuen, N, Zhao, Y, Min, J, Frasor, J, Katzenellenbogen, B.S, Griffin, P.R, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-06-06 | | Release date: | 2016-11-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Full antagonism of the estrogen receptor without a prototypical ligand side chain.
Nat. Chem. Biol., 13, 2017
|
|
5KCT
 
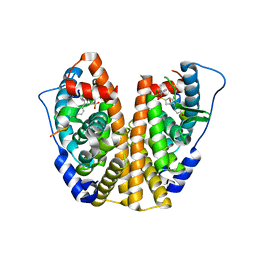 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with an N-ethyl, 4-chlorobenzyl OBHS-N derivative | | Descriptor: | (1R,2S,4R)-N-(4-chlorophenyl)-N-ethyl-5,6-bis(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonamide, (1S,2R,4S)-N-(4-chlorophenyl)-N-ethyl-5,6-bis(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonamide, Estrogen receptor, ... | | Authors: | Nwachukwu, J.C, Srinivasan, S, Bruno, N.E, Dharmarajan, V, Goswami, D, Kastrati, I, Novick, S, Nowak, J, Zhou, H.B, Boonmuen, N, Zhao, Y, Min, J, Frasor, J, Katzenellenbogen, B.S, Griffin, P.R, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-06-07 | | Release date: | 2016-11-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Full antagonism of the estrogen receptor without a prototypical ligand side chain.
Nat. Chem. Biol., 13, 2017
|
|
5E15
 
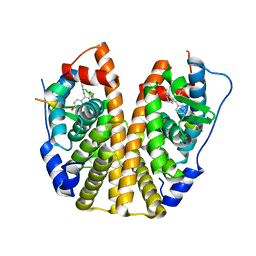 | | Crystal Structure of the ER-alpha Ligand-binding Domain in Complex with the Cyclofenil Derivative 4,4'-{[4-(2-hydroxyethyl)cyclohexylidene]methanediyl}diphenol | | Descriptor: | 4,4'-({4-[2-(4-fluorobutoxy)ethyl]cyclohexylidene}methanediyl)diphenol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | Deposit date: | 2015-09-29 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
5RE4
 
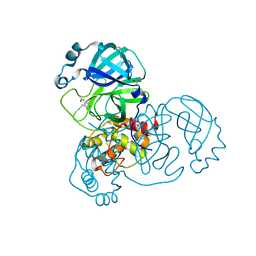 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 main protease in complex with Z1129283193 | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, N-(4-methylpyridin-3-yl)acetamide | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-03-15 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
5REL
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 main protease in complex with PCM-0102340 | | Descriptor: | 1-{4-[(3-methylphenyl)methyl]piperazin-1-yl}ethan-1-one, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-03-15 | | Release date: | 2020-03-25 | | Last modified: | 2021-02-24 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
5RF1
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 main protease in complex with NCL-00023830 | | Descriptor: | 3C-like proteinase, 4-bromobenzene-1-sulfonamide, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-03-15 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
5RFF
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 main protease in complex with PCM-0102704 | | Descriptor: | 1-{4-[(4-chlorophenyl)sulfonyl]piperazin-1-yl}ethan-1-one, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-03-15 | | Release date: | 2020-03-25 | | Last modified: | 2021-02-24 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
5RFV
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 main protease in complex with PCM-0102306 | | Descriptor: | 1-[4-(thiophene-2-carbonyl)piperazin-1-yl]ethan-1-one, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-03-15 | | Release date: | 2020-03-25 | | Last modified: | 2021-02-24 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
5KEQ
 
 | | High resolution cryo-EM maps of Human papillomavirus 16 reveal L2 location and heparin-induced conformational changes | | Descriptor: | Major capsid protein L1 | | Authors: | Guan, J, Bywaters, S.M, Brendle, S.A, Ashley, R.E, Makhov, A.M, Conway, J.F, Christensen, N.D, Hafenstein, S. | | Deposit date: | 2016-06-10 | | Release date: | 2017-01-25 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Cryoelectron Microscopy Maps of Human Papillomavirus 16 Reveal L2 Densities and Heparin Binding Site.
Structure, 25, 2017
|
|
5RHD
 
 | | PanDDA analysis group deposition SARS-CoV-2 main protease fragment screen -- Crystal Structure of SARS-CoV-2 main protease in complex with SF013 (Mpro-x2193) | | Descriptor: | 1-[4-(methylsulfonyl)phenyl]piperazine, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, Keeley, A, Keseru, G.M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-05-16 | | Release date: | 2020-06-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
7DKP
 
 | |
2BOJ
 
 | | crystal Structure of pseudomonas aeruginosa lectin (PA-IIL) complexed with methyl-B-D-Arabinopyranoside | | Descriptor: | CALCIUM ION, PSEUDOMONAS AERUGINOSA LECTIN II, SULFATE ION, ... | | Authors: | Sabin, C.D, Mitchell, E.P, Wimmerova, M, Imberty, A. | | Deposit date: | 2005-04-12 | | Release date: | 2006-02-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Binding of Different Monosaccharides by Lectin Pa-Iil from Pseudoman Aeruginosa: Thermodynamics Data Correlated with X-Ray Structures.
FEBS Lett., 580, 2006
|
|
7VSO
 
 | | Serial Femtosecond Crystallography (SFX) of Ground State Bacteriorhodopsin Crystallized from Bicelles in Complex with HAD16 Determined Using 7-keV X-ray Free Electron Laser (XFEL) at SACLA | | Descriptor: | 2-[[(2R)-2-(3-bromanyl-5-iodanyl-phenyl)carbonyloxy-3-tetradecanoyloxy-propoxy]-oxidanyl-phosphoryl]oxyethyl-trimethyl-azanium, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, Bacteriorhodopsin, ... | | Authors: | Mizohata, E, Nakane, T, Hanashima, S. | | Deposit date: | 2021-10-27 | | Release date: | 2022-02-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Heavy Atom Detergent/Lipid Combined X-ray Crystallography for Elucidating the Structure-Function Relationships of Membrane Proteins.
Membranes (Basel), 11, 2021
|
|
5RE9
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 main protease in complex with Z2856434836 | | Descriptor: | 2-(4-methylphenoxy)-1-(4-methylpiperazin-4-ium-1-yl)ethanone, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-03-15 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
5REP
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 main protease in complex with PCM-0102201 | | Descriptor: | 1-{4-[(2,6-difluorophenyl)sulfonyl]piperazin-1-yl}ethan-1-one, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-03-15 | | Release date: | 2020-03-25 | | Last modified: | 2021-02-24 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
1WS0
 
 | |
5RF9
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 main protease in complex with Z217038356 | | Descriptor: | 1-[(2~{S})-2-methylmorpholin-4-yl]-2-pyrazol-1-yl-ethanone, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-03-15 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
5RFL
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 main protease in complex with PCM-0102389 | | Descriptor: | 1-acetyl-N-(2-hydroxyphenyl)piperidine-4-carboxamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-03-15 | | Release date: | 2020-03-25 | | Last modified: | 2021-02-24 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
5RFZ
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 main protease in complex with PCM-0102274 | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, N-(2-chloropyridin-3-yl)acetamide | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-03-15 | | Release date: | 2020-03-25 | | Last modified: | 2021-02-24 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
4HGY
 
 | | Structure of the CcbJ Methyltransferase from Streptomyces caelestis | | Descriptor: | 1,2-ETHANEDIOL, CcbJ, SULFATE ION | | Authors: | Bauer, J.A, Ondrovicova, G, Kutejova, E, Janata, J. | | Deposit date: | 2012-10-09 | | Release date: | 2013-10-30 | | Last modified: | 2014-04-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure and possible mechanism of the CcbJ methyltransferase from Streptomyces caelestis.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
5RGN
 
 | | PanDDA analysis group deposition SARS-CoV-2 main protease fragment screen -- Crystal Structure of SARS-CoV-2 main protease in complex with PCM-0102759 (Mpro-x0731) | | Descriptor: | 1-{4-[(4-methylphenyl)sulfonyl]piperazin-1-yl}ethan-1-one, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-04-07 | | Release date: | 2020-04-15 | | Last modified: | 2021-02-24 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
