2X57
 
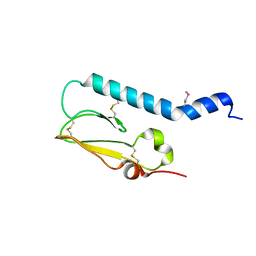 | | Crystal structure of the extracellular domain of human Vasoactive intestinal polypeptide receptor 2 | | Descriptor: | VASOACTIVE INTESTINAL POLYPEPTIDE RECEPTOR 2 | | Authors: | Pike, A.C.W, Barr, A.J, Quigley, A, Burgess Brown, N, de Riso, A, Bullock, A, Berridge, G, Muniz, J.R.C, Chaikaud, A, Vollmar, M, Krojer, T, Ugochukwu, E, von Delft, F, Edwards, A, Arrowsmith, C.H, Weigelt, J, Bountra, C, Carpenter, E.P. | | Deposit date: | 2010-02-05 | | Release date: | 2010-03-09 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of the Extracellular Domain of Human Vasoactive Intestinal Polypeptide Receptor 2
To be Published
|
|
6GFX
 
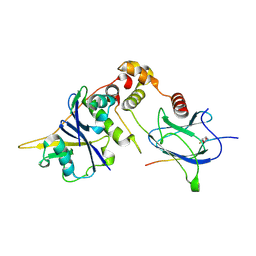 | | pVHL:EloB:EloC in complex with modified HIF-1a CODD peptide containing (3R,4S)-3-fluoro-4-hydroxyproline (ligand 13a) | | Descriptor: | Elongin-B, Elongin-C, FLUORINATED HYPOXIA-INDUCIBLE FACTOR 1 ALPHA PEPTIDE, ... | | Authors: | Castro, G.V, Testa, A, Ciulli, A. | | Deposit date: | 2018-05-02 | | Release date: | 2018-07-11 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | 3-Fluoro-4-hydroxyprolines: Synthesis, Conformational Analysis, and Stereoselective Recognition by the VHL E3 Ubiquitin Ligase for Targeted Protein Degradation.
J. Am. Chem. Soc., 140, 2018
|
|
6GFZ
 
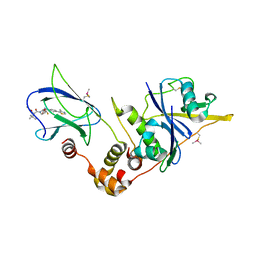 | | pVHL:EloB:EloC in complex with modified VH032 containing (3S,4S)-3-fluoro-4-hydroxyproline (ligand 14b) | | Descriptor: | (2~{R},3~{S},4~{S})-1-[(2~{S})-2-acetamido-3,3-dimethyl-butanoyl]-3-fluoranyl-~{N}-[[4-(4-methyl-1,3-thiazol-5-yl)phenyl]methyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Elongin-B, Elongin-C, ... | | Authors: | Gadd, M.S, Testa, A, Ciulli, A. | | Deposit date: | 2018-05-02 | | Release date: | 2018-07-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | 3-Fluoro-4-hydroxyprolines: Synthesis, Conformational Analysis, and Stereoselective Recognition by the VHL E3 Ubiquitin Ligase for Targeted Protein Degradation.
J. Am. Chem. Soc., 140, 2018
|
|
2CBQ
 
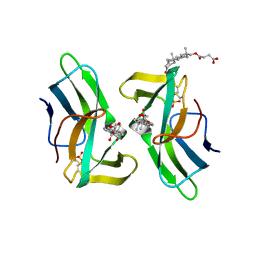 | | Crystal structure of the neocarzinostatin 1Tes15 mutant bound to testosterone hemisuccinate. | | Descriptor: | NEOCARZINOSTATIN, SULFATE ION, TESTOSTERONE HEMISUCCINATE | | Authors: | Drevelle, A, Graille, M, Heyd, B, Sorel, I, Ulryck, N, Pecorari, F, Desmadril, M, Van Tilbeurgh, H, Minard, P. | | Deposit date: | 2006-01-06 | | Release date: | 2006-03-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of in Vitro Evolved Binding Sites on Neocarzinostatin Scaffold Reveal Unanticipated Evolutionary Pathways.
J.Mol.Biol., 358, 2006
|
|
2CBO
 
 | | Crystal structure of the neocarzinostatin 3Tes24 mutant bound to testosterone hemisuccinate. | | Descriptor: | NEOCARZINOSTATIN, SULFATE ION, TESTOSTERONE HEMISUCCINATE | | Authors: | Drevelle, A, Graille, M, Heyd, B, Sorel, I, Ulryck, N, Pecorari, F, Desmadril, M, van Tilbeurgh, H, Minard, P. | | Deposit date: | 2006-01-06 | | Release date: | 2006-03-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of in Vitro Evolved Binding Sites on Neocarzinostatin Scaffold Reveal Unanticipated Evolutionary Pathways.
J.Mol.Biol., 358, 2006
|
|
2CBT
 
 | | Crystal structure of the neocarzinostatin 4Tes1 mutant bound testosterone hemisuccinate. | | Descriptor: | NEOCARZINOSTATIN, TESTOSTERONE HEMISUCCINATE | | Authors: | Drevelle, A, Graille, M, Heyd, B, Sorel, I, Ulryck, N, Pecorari, F, Desmadril, M, Van Tilbeurgh, H, Minard, P. | | Deposit date: | 2006-01-06 | | Release date: | 2006-03-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of in Vitro Evolved Binding Sites on Neocarzinostatin Scaffold Reveal Unanticipated Evolutionary Pathways.
J.Mol.Biol., 358, 2006
|
|
5L94
 
 | |
3NBR
 
 | | Crystal Structure of Ketosteroid Isomerase D38NP39GD99N from Pseudomonas Testosteroni (tKSI) with 4-Androstene-3,17-dione Bound | | Descriptor: | 4-ANDROSTENE-3-17-DIONE, SULFATE ION, Steroid Delta-isomerase | | Authors: | Gonzalez, A, Tsai, Y, Schwans, J, Ruben, E, Sunden, F, Herschlag, D. | | Deposit date: | 2010-06-03 | | Release date: | 2011-11-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal Structure of Ketosteroid Isomerase D38NP39GD99N from Pseudomonas Testosteroni (tKSI) with 4-Androstene-3,17-dione Bound
To be Published
|
|
3NXJ
 
 | | Crystal Structure of Ketosteroid Isomerase D99N from Pseudomonas Testosteroni (tKSI) | | Descriptor: | SULFATE ION, Steroid Delta-isomerase | | Authors: | Gonzalez, A, Tsai, Y, Schwans, J, Sunden, F, Herschlag, D. | | Deposit date: | 2010-07-13 | | Release date: | 2011-08-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.966 Å) | | Cite: | Crystal Structure of Ketosteroid Isomerase
D99N from Pseudomonas Testosteroni (tKSI)
To be Published
|
|
3NUV
 
 | | Crystal structure of ketosteroid isomerase D38ND99N from Pseudomonas testosteroni (tKSI) with 4-Androstene-3,17-dione Bound | | Descriptor: | 4-ANDROSTENE-3-17-DIONE, SULFATE ION, Steroid Delta-isomerase | | Authors: | Gonzalez, A, Tsai, Y, Schwans, J, Sunden, F, Herschlag, D. | | Deposit date: | 2010-07-07 | | Release date: | 2011-11-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal Structure of Ketosteroid Isomerase
D38ND99N from Pseudomonas testosteroni (tKSI) with 4-Androstene-3,17-dione Bound
TO BE PUBLISHED
|
|
3OV4
 
 | | Crystal Structure of Ketosteroid Isomerase P39GV40GS42G from Pseudomonas Testosteroni (tKSI) bound to Equilenin | | Descriptor: | EQUILENIN, SULFATE ION, Steroid Delta-isomerase | | Authors: | Gonzalez, A, Tsai, Y, Schwans, J, Sunden, F, Herschlag, D. | | Deposit date: | 2010-09-15 | | Release date: | 2011-10-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal Structure of Ketosteroid Isomerase
P39GV40GS42G from Pseudomonas Testosteroni (tKSI)
To be Published
|
|
3MKI
 
 | | Crystal Structure of Ketosteroid Isomerase D38ED99N from Pseudomonas Testosteroni (tKSI) | | Descriptor: | GLYCEROL, SULFATE ION, Steroid Delta-Isomerase | | Authors: | Gonzalez, A, Tsai, Y, Schwans, J, Sunden, F, Herschlag, D. | | Deposit date: | 2010-04-14 | | Release date: | 2011-11-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Ketosteroid Isomerase D38E,D99N from Pseudomonas Testosteroni (tKSI)
To be Published
|
|
3M8C
 
 | | Crystal Structure of Ketosteroid Isomerase D99N from Pseudomonas Testosteroni (tKSI) with Equilenin Bound | | Descriptor: | EQUILENIN, GLYCEROL, SULFATE ION, ... | | Authors: | Gonzalez, A, Tsai, Y, Schwans, J, Sunden, F, Herschlag, D. | | Deposit date: | 2010-03-17 | | Release date: | 2010-04-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Ketosteroid Isomerase D99N from Pseudomonas Testosteroni (tKSI) with Equilenin Bound
TO BE PUBLISHED
|
|
3NM2
 
 | | Crystal Structure of Ketosteroid Isomerase D38EP39GV40GS42G from Pseudomonas Testosteroni (tKSI) | | Descriptor: | SULFATE ION, Steroid Delta-isomerase | | Authors: | Gonzalez, A, Tsai, Y, Schwans, J, Sunden, F, Herschlag, D. | | Deposit date: | 2010-06-21 | | Release date: | 2011-11-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.887 Å) | | Cite: | Crystal Structure of Ketosteroid Isomerase D38EP39GV40GS42G from Pseudomonas Testosteroni (tKSI)
To be Published
|
|
3MYT
 
 | | Crystal structure of Ketosteroid Isomerase D38HD99N from Pseudomonas testosteroni (tKSI) | | Descriptor: | EQUILENIN, GLYCEROL, SULFATE ION, ... | | Authors: | Gonzalez, A, Tsai, Y, Schwans, J, Sunden, F, Herschlag, D. | | Deposit date: | 2010-05-11 | | Release date: | 2011-11-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.961 Å) | | Cite: | Crystal Structure of Ketosteroid Isomerase D38HD99N from Pseudomonas Testosteroni (tKSI)
To be Published
|
|
3NHX
 
 | | Crystal Structure of Ketosteroid Isomerase D99N from Pseudomonas Testosteroni (tKSI) with 4-Androstene-3,17-dione Bound | | Descriptor: | 4-ANDROSTENE-3-17-DIONE, SULFATE ION, Steroid Delta-isomerase | | Authors: | Gonzalez, A, Tsai, Y, Schwans, J, Ruben, E, Sunden, F, Herschlag, D. | | Deposit date: | 2010-06-14 | | Release date: | 2011-11-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Crystal Structure of Ketosteroid Isomerase D99N from Pseudomonas Testosteroni (tKSI) with 4-Androstene-3,17-dione Bound
To be Published
|
|
2MJI
 
 | | HIFABP_Ketorolac_complex | | Descriptor: | (1R)-5-benzoyl-2,3-dihydro-1H-pyrrolizine-1-carboxylic acid, Fatty acid-binding protein, intestinal | | Authors: | Patil, R, Laguerre, A, Wielens, J, Headey, S, Williams, M, Mohanty, B, Porter, C, Scanlon, M. | | Deposit date: | 2014-01-09 | | Release date: | 2014-10-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Characterization of two distinct modes of drug binding to human intestinal Fatty Acid binding protein.
Acs Chem.Biol., 9, 2014
|
|
2OC2
 
 | | Structure of testis ACE with RXPA380 | | Descriptor: | Angiotensin-converting enzyme, somatic isoform, CHLORIDE ION, ... | | Authors: | Corradi, H.R, Anthony, C.S, Schwager, S.L, Redelinghuys, P, Georgiadis, D, Dive, V, Acharya, K.R, Sturrock, E.D. | | Deposit date: | 2006-12-20 | | Release date: | 2007-05-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The structure of testis angiotensin-converting enzyme in complex with the C domain-specific inhibitor RXPA380.
Biochemistry, 46, 2007
|
|
4OED
 
 | | Crystal structure of AR-LBD bound with co-regulator peptide | | Descriptor: | 5-ALPHA-DIHYDROTESTOSTERONE, Androgen receptor, Protein BUD31 homolog, ... | | Authors: | Liu, J.S, Hsu, C.L, Wu, W.G. | | Deposit date: | 2014-01-13 | | Release date: | 2014-08-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Identification of a new androgen receptor (AR) co-regulator BUD31 and related peptides to suppress wild-type and mutated AR-mediated prostate cancer growth via peptide screening and X-ray structure analysis.
Mol Oncol, 8, 2014
|
|
4OEA
 
 | | Crystal structure of AR-LBD | | Descriptor: | 5-ALPHA-DIHYDROTESTOSTERONE, Androgen receptor | | Authors: | Liu, J.S, Hsu, C.L, Wu, W.G. | | Deposit date: | 2014-01-12 | | Release date: | 2014-08-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Identification of a new androgen receptor (AR) co-regulator BUD31 and related peptides to suppress wild-type and mutated AR-mediated prostate cancer growth via peptide screening and X-ray structure analysis.
Mol Oncol, 8, 2014
|
|
4OFR
 
 | | Crystal structure of AR-LBD bound with co-regulator peptide | | Descriptor: | 5-ALPHA-DIHYDROTESTOSTERONE, Androgen receptor, co-regulator peptide | | Authors: | Liu, J.S, Hsu, C.L, Wu, W.G. | | Deposit date: | 2014-01-15 | | Release date: | 2014-08-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Identification of a new androgen receptor (AR) co-regulator BUD31 and related peptides to suppress wild-type and mutated AR-mediated prostate cancer growth via peptide screening and X-ray structure analysis.
Mol Oncol, 8, 2014
|
|
4OFU
 
 | | Crystal structure of AR-LBD bound with co-regulator peptide | | Descriptor: | 5-ALPHA-DIHYDROTESTOSTERONE, Androgen receptor, co-regulator peptide | | Authors: | Liu, J.S, Hsu, C.L, Wu, W.G. | | Deposit date: | 2014-01-15 | | Release date: | 2014-08-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Identification of a new androgen receptor (AR) co-regulator BUD31 and related peptides to suppress wild-type and mutated AR-mediated prostate cancer growth via peptide screening and X-ray structure analysis.
Mol Oncol, 8, 2014
|
|
4OEZ
 
 | | Crystal structure of AR-LBD bound with co-regulator peptide | | Descriptor: | 5-ALPHA-DIHYDROTESTOSTERONE, Androgen receptor, SULFATE ION, ... | | Authors: | Liu, J.S, Hsu, C.L, Wu, W.G. | | Deposit date: | 2014-01-14 | | Release date: | 2014-08-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification of a new androgen receptor (AR) co-regulator BUD31 and related peptides to suppress wild-type and mutated AR-mediated prostate cancer growth via peptide screening and X-ray structure analysis.
Mol Oncol, 8, 2014
|
|
4OEY
 
 | | Crystal structure of AR-LBD bound with co-regulator peptide | | Descriptor: | 5-ALPHA-DIHYDROTESTOSTERONE, Androgen receptor, SULFATE ION, ... | | Authors: | Liu, J.S, Hsu, C.L, Wu, W.G. | | Deposit date: | 2014-01-14 | | Release date: | 2014-08-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Identification of a new androgen receptor (AR) co-regulator BUD31 and related peptides to suppress wild-type and mutated AR-mediated prostate cancer growth via peptide screening and X-ray structure analysis.
Mol Oncol, 8, 2014
|
|
2QLY
 
 | | Crystral Structure of the N-terminal Subunit of Human Maltase-Glucoamylase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Sim, L, Rose, D.R. | | Deposit date: | 2007-07-13 | | Release date: | 2008-01-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Human intestinal maltase-glucoamylase: crystal structure of the N-terminal catalytic subunit and basis of inhibition and substrate specificity
J.Mol.Biol., 375, 2008
|
|
