4C16
 
 | | E-selectin lectin, EGF-like and two SCR domains complexed with glycomimetic antagonist | | Descriptor: | (1R,2R,3S)-3-methylcyclohexane-1,2-diol, (S)-CYCLOHEXYL LACTIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Preston, R.C, Jakob, R.P, Binder, F.P.C, Sager, C.P, Ernst, B, Maier, T. | | Deposit date: | 2013-08-09 | | Release date: | 2014-08-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | E-Selectin Ligand Complexes Adopt an Extended High-Affinity Conformation.
J.Mol.Cell.Biol., 8, 2016
|
|
7P49
 
 | | HLA-E*01:03 in complex with Mtb14 | | Descriptor: | Beta-2-microglobulin, GLYCEROL, HLA class I histocompatibility antigen, ... | | Authors: | Walters, L.C, Gillespie, G.M. | | Deposit date: | 2021-07-10 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Primary and secondary functions of HLA-E are determined by stability and conformation of the peptide-bound complexes.
Cell Rep, 39, 2022
|
|
4HR7
 
 | | Crystal Structure of Biotin Carboxyl Carrier Protein-Biotin Carboxylase Complex from E.coli | | Descriptor: | 1,2-ETHANEDIOL, Biotin carboxyl carrier protein of acetyl-CoA carboxylase, Biotin carboxylase, ... | | Authors: | Broussard, T.C, Kobe, M.J, Pakhomova, S, Neau, D.B, Price, A.E, Champion, T.S, Waldrop, G.L. | | Deposit date: | 2012-10-26 | | Release date: | 2013-03-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.495 Å) | | Cite: | The three-dimensional structure of the biotin carboxylase-biotin carboxyl carrier protein complex of E. coli acetyl-CoA carboxylase.
Structure, 21, 2013
|
|
7P4B
 
 | | HLA-E*01:03 in complex with IL9 | | Descriptor: | Beta-2-microglobulin, ESAT-6-like protein EsxH, GLYCEROL, ... | | Authors: | Walters, L.C, Gillespie, G.M. | | Deposit date: | 2021-07-10 | | Release date: | 2022-07-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Primary and secondary functions of HLA-E are determined by stability and conformation of the peptide-bound complexes.
Cell Rep, 39, 2022
|
|
8BYX
 
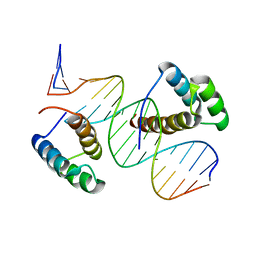 | |
3TTQ
 
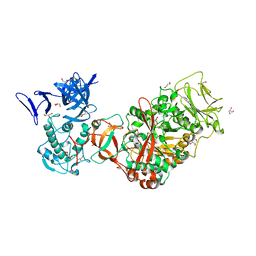 | | Crystal structure of Leuconostoc mesenteroides NRRL B-1299 N-terminally truncated dextransucrase DSR-E in orthorhombic apo-form at 1.9 angstrom resolution | | Descriptor: | CALCIUM ION, Dextransucrase, GLYCEROL, ... | | Authors: | Brison, Y, Pijning, T, Fabre, E, Mourey, L, Morel, S, Potocki-Veronese, G, Monsan, P, Remaud-Simeon, M, Dijkstra, B.W, Tranier, S. | | Deposit date: | 2011-09-15 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Functional and structural characterization of alpha-(1-2) branching sucrase derived from DSR-E glucansucrase
J.Biol.Chem., 287, 2012
|
|
3KMH
 
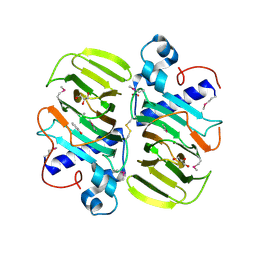 | |
7UWH
 
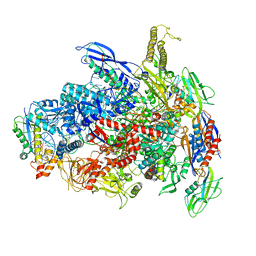 | | CryoEM Structure of E. coli Transcription-Coupled Ribonucleotide Excision Repair (TC-RER) complex bound to ribonucleotide substrate | | Descriptor: | DNA (59-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Hao, Z.T, Grower, M, Bharati, B, Proshkin, S, Epshtein, V, Svetlov, V, Nudler, E, Shamovsky, I. | | Deposit date: | 2022-05-03 | | Release date: | 2023-05-31 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | RNA polymerase drives ribonucleotide excision DNA repair in E. coli.
Cell, 186, 2023
|
|
7UWE
 
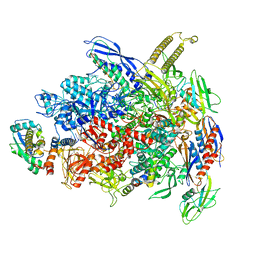 | | CryoEM Structure of E. coli Transcription-Coupled Ribonucleotide Excision Repair (TC-RER) complex | | Descriptor: | DNA (29-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Hao, Z.T, Grower, M, Bharati, B, Proshkin, S, Epshtein, V, Svetlov, V, Nudler, E, Shamovsky, I. | | Deposit date: | 2022-05-03 | | Release date: | 2023-05-31 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | RNA polymerase drives ribonucleotide excision DNA repair in E. coli.
Cell, 186, 2023
|
|
8ELM
 
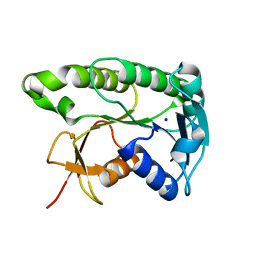 | | Apo human biliverdin reductase beta (293K) | | Descriptor: | Flavin reductase (NADPH), SODIUM ION | | Authors: | McLeod, M.J, Eisenmesser, E.Z, Lee, E, Thorne, R.E. | | Deposit date: | 2022-09-26 | | Release date: | 2023-09-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Identifying structural and dynamic changes during the Biliverdin Reductase B catalytic cycle.
Front Mol Biosci, 10, 2023
|
|
8R7F
 
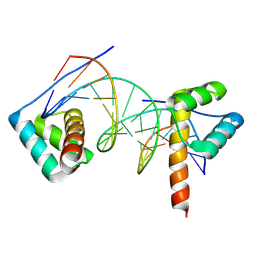 | | Transcription factor BARHL2 homodimer with spacing two bp | | Descriptor: | BarH-like 2 homeobox protein, DNA (5'-D(*CP*TP*AP*AP*AP*CP*GP*GP*GP*CP*AP*AP*TP*TP*AP*G)-3'), DNA (5'-D(*CP*TP*AP*AP*TP*TP*GP*CP*CP*CP*GP*TP*TP*TP*AP*G)-3') | | Authors: | Morgunova, E, Popov, A, Yin, Y, Taipale, J. | | Deposit date: | 2023-11-24 | | Release date: | 2024-12-04 | | Last modified: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | DNA-guided transcription factor interactions extend human gene regulatory code.
Nature, 641, 2025
|
|
8R7Z
 
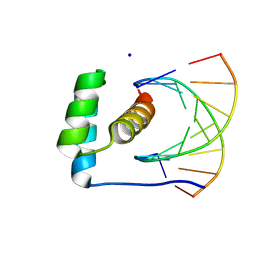 | | transcription factor BARHL2 homodimer with spacing four bp | | Descriptor: | BarH-like 2 homeobox protein, DNA (5'-D(P*AP*CP*CP*GP*TP*TP*TP*AP*G)-3'), DNA (5'-D(P*CP*TP*AP*AP*AP*CP*GP*GP*T)-3'), ... | | Authors: | Morgunova, E, Popov, A, Yin, Y, Taipale, J. | | Deposit date: | 2023-11-27 | | Release date: | 2024-12-04 | | Last modified: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (3.26 Å) | | Cite: | DNA-guided transcription factor interactions extend human gene regulatory code.
Nature, 641, 2025
|
|
6HPB
 
 | |
8ELL
 
 | | Apo human biliverdin reductase beta (cryogenic) | | Descriptor: | Flavin reductase (NADPH), SODIUM ION | | Authors: | McLeod, M.J, Eisenmesser, E.Z, Lee, E, Thorne, R.E. | | Deposit date: | 2022-09-26 | | Release date: | 2023-09-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Identifying structural and dynamic changes during the Biliverdin Reductase B catalytic cycle.
Front Mol Biosci, 10, 2023
|
|
3MPB
 
 | |
8V5P
 
 | |
8JJ6
 
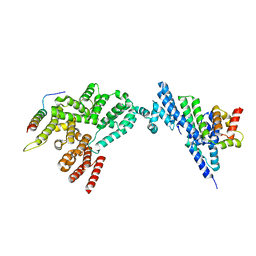 | | Structure of the NELF-BCE complex | | Descriptor: | NELF-E, Negative elongation factor B, Negative elongation factor complex member C/D | | Authors: | Wang, Z, Cao, Y, Qin, Y. | | Deposit date: | 2023-05-29 | | Release date: | 2023-08-30 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Structural basis of the human negative elongation factor NELF-B/C/E ternary complex.
Biochem.Biophys.Res.Commun., 677, 2023
|
|
8V5Q
 
 | | Varicella Zoster Virus (VZV) glycoprotein E (gE) gI binding domain in complex with human Fab 1E3 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Envelope glycoprotein E, Fab 1E3 Heavy Chain, ... | | Authors: | Holzapfel, G, Seraj, N, Harshbarger, W. | | Deposit date: | 2023-11-30 | | Release date: | 2024-12-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of the Varicella Zoster Virus Glycoprotein E and Epitope Mapping of Vaccine-Elicited Antibodies.
Vaccines (Basel), 12, 2024
|
|
8BZM
 
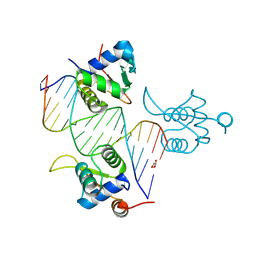 | | FOXK1-ELF1-heterodimer bound to DNA | | Descriptor: | DNA, ETS-related transcription factor Elf-1, Forkhead box protein K1, ... | | Authors: | Morgunova, E, Popov, A, Yin, Y, Taipale, J. | | Deposit date: | 2022-12-15 | | Release date: | 2023-12-27 | | Last modified: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | DNA-guided transcription factor interactions extend human gene regulatory code.
Nature, 2025
|
|
2GNN
 
 | | Crystal Structure of the Orf Virus NZ2 Variant of VEGF-E | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, BENZAMIDINE, ... | | Authors: | Prota, A.E, Pieren, M, Wagner, A, Kostrewa, D, Winkler, F.K, Ballmer-Hofer, K. | | Deposit date: | 2006-04-10 | | Release date: | 2006-05-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the Orf virus NZ2 variant of vascular endothelial growth factor-E. Implications for receptor specificity.
J.Biol.Chem., 281, 2006
|
|
7X7R
 
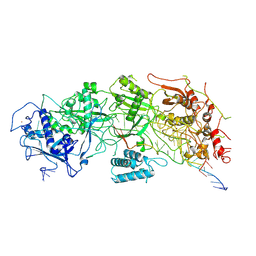 | | Cryo-EM structure of a bacterial protein | | Descriptor: | RAMP superfamily protein, RNA (36-MER), RNA (5'-R(P*AP*GP*UP*CP*CP*GP*GP*GP*GP*CP*AP*GP*AP*AP*AP*AP*UP*UP*GP*G)-3'), ... | | Authors: | Yu, G, Wang, X, Deng, Z, Zhang, H. | | Deposit date: | 2022-03-10 | | Release date: | 2022-11-16 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure and function of a bacterial type III-E CRISPR-Cas7-11 complex.
Nat Microbiol, 7, 2022
|
|
7XC7
 
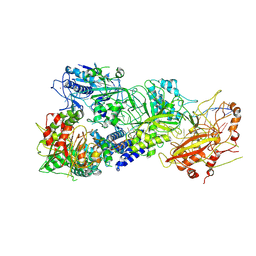 | | Cryo-EM structure of a bacterial protein complex | | Descriptor: | CHAT domain protein, RAMP superfamily protein, RNA (33-MER), ... | | Authors: | Yu, G, Wang, X, Deng, Z, Zhang, H. | | Deposit date: | 2022-03-23 | | Release date: | 2022-11-16 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure and function of a bacterial type III-E CRISPR-Cas7-11 complex.
Nat Microbiol, 7, 2022
|
|
7X8A
 
 | | Cryo-EM structure of a bacterial protein complex | | Descriptor: | CHAT domain protein, RAMP superfamily protein, RNA (33-MER), ... | | Authors: | Yu, G, Wang, X, Deng, Z, Zhang, H. | | Deposit date: | 2022-03-11 | | Release date: | 2022-11-16 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structure and function of a bacterial type III-E CRISPR-Cas7-11 complex.
Nat Microbiol, 7, 2022
|
|
7X7A
 
 | | Cryo-EM structure of SbCas7-11 in complex with crRNA and target RNA | | Descriptor: | RAMP superfamily protein, RNA (33-MER), ZINC ION | | Authors: | Yu, G, Wang, X, Deng, Z, Zhang, H. | | Deposit date: | 2022-03-09 | | Release date: | 2022-11-16 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure and function of a bacterial type III-E CRISPR-Cas7-11 complex.
Nat Microbiol, 7, 2022
|
|
9AZS
 
 | |
