1SC5
 
 | | Sigma-28(FliA)/FlgM complex | | Descriptor: | RNA polymerase sigma factor FliA, anti-sigma factor FlgM | | Authors: | Sorenson, M.K, Ray, S.S, Darst, S.A. | | Deposit date: | 2004-02-11 | | Release date: | 2004-04-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.26 Å) | | Cite: | Crystal structure of the flagellar sigma/anti-sigma complex sigma(28)/FlgM reveals an intact sigma factor in an inactive conformation.
Mol.Cell, 14, 2004
|
|
7RNA
 
 | |
4RNA
 
 | | Crystal structure of PLpro from Middle East Respiratory Syndrome (MERS) coronavirus | | Descriptor: | PHOSPHATE ION, ZINC ION, papain-like protease | | Authors: | Lei, H, Santarsiero, B.D, Lee, H, Johnson, M.E. | | Deposit date: | 2014-10-23 | | Release date: | 2015-03-25 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.794 Å) | | Cite: | Inhibitor Recognition Specificity of MERS-CoV Papain-like Protease May Differ from That of SARS-CoV.
Acs Chem.Biol., 10, 2015
|
|
5HPO
 
 | | Cycloalternan-forming enzyme from Listeria monocytogenes in complex with maltopentaose | | Descriptor: | CALCIUM ION, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Halavaty, A.S, Light, S.H, Minasov, G, Winsor, J, Grimshaw, S, Shuvalova, L, Peterson, S, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-01-20 | | Release date: | 2017-01-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Transferase Versus Hydrolase: The Role of Conformational Flexibility in Reaction Specificity.
Structure, 25, 2017
|
|
5HVQ
 
 | | Alternative model of the MAGE-G1 NSE-1 complex | | Descriptor: | Melanoma-associated antigen G1, Non-structural maintenance of chromosomes element 1 homolog, ZINC ION | | Authors: | Newman, J.A, Cooper, C.D.O, Roos, A.K, Aitkenhead, H, Oppermann, U.C.T, Cho, H.J, Osman, R, Gileadi, O. | | Deposit date: | 2016-01-28 | | Release date: | 2016-10-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.923 Å) | | Cite: | Structures of Two Melanoma-Associated Antigens Suggest Allosteric Regulation of Effector Binding.
Plos One, 11, 2016
|
|
2L6I
 
 | |
1M9S
 
 | | Crystal structure of Internalin B (InlB), a Listeria monocytogenes virulence protein containing SH3-like domains. | | Descriptor: | Internalin B, SULFATE ION, TERBIUM(III) ION | | Authors: | Marino, M, Banerjee, M, Jonquieres, R, Cossart, P, Ghosh, P. | | Deposit date: | 2002-07-29 | | Release date: | 2002-11-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | GW domains of the Listeria monocytogenes invasion protein InlB are SH3-like and mediate binding to host ligands
Embo J., 21, 2002
|
|
2MAO
 
 | |
6MHT
 
 | |
3ARG
 
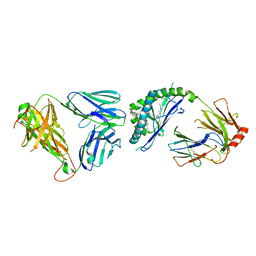 | | Ternary crystal structure of the mouse NKT TCR-CD1d-alpha-glucosylceramide(C20:2) | | Descriptor: | (11E,14E)-N-[(2S,3S,4R)-1-(alpha-D-glucopyranosyloxy)-3,4-dihydroxyoctadecan-2-yl]icosa-11,14-dienamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wun, K.S, Rossjohn, J. | | Deposit date: | 2010-11-27 | | Release date: | 2011-03-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A molecular basis for the exquisite CD1d-restricted antigen specificity and functional responses of natural killer T cells
Immunity, 34, 2011
|
|
3ARB
 
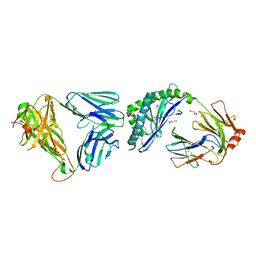 | | Ternary crystal structure of the NKT TCR-CD1d-alpha-galactosylceramide analogue-OCH | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d1, ... | | Authors: | Wun, K.S, Rossjohn, J. | | Deposit date: | 2010-11-26 | | Release date: | 2011-03-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A molecular basis for the exquisite CD1d-restricted antigen specificity and functional responses of natural killer T cells
Immunity, 34, 2011
|
|
3ARD
 
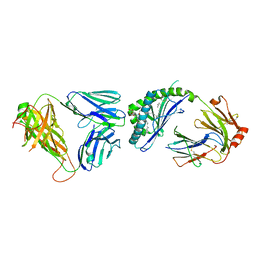 | | Ternary crystal structure of the mouse NKT TCR-CD1d-3'deoxy-alpha-galactosylceramide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d1, Beta-2-microglobulin, ... | | Authors: | Wun, K.S, Rossjohn, J. | | Deposit date: | 2010-11-27 | | Release date: | 2011-03-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | A molecular basis for the exquisite CD1d-restricted antigen specificity and functional responses of natural killer T cells
Immunity, 34, 2011
|
|
3ARE
 
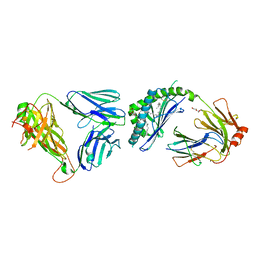 | | Ternary crystal structure of the mouse NKT TCR-CD1d-4'deoxy-alpha-galactosylceramide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d1, ... | | Authors: | Wun, K.S, Rossjohn, J. | | Deposit date: | 2010-11-27 | | Release date: | 2011-03-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A molecular basis for the exquisite CD1d-restricted antigen specificity and functional responses of natural killer T cells
Immunity, 34, 2011
|
|
3ARF
 
 | | Ternary crystal structure of the mouse NKT TCR-CD1d-C20:2 | | Descriptor: | (11Z,14E)-N-[(2S,3S,4R)-1-(alpha-D-galactopyranosyloxy)-3,4-dihydroxyoctadecan-2-yl]icosa-11,14-dienamide, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d1, ... | | Authors: | Wun, K.S, Rossjohn, J. | | Deposit date: | 2010-11-27 | | Release date: | 2011-03-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A molecular basis for the exquisite CD1d-restricted antigen specificity and functional responses of natural killer T cells
Immunity, 34, 2011
|
|
8OV6
 
 | | Ternary structure of intramolecular bivalent glue degrader IBG1 bound to BRD4 and DCAF16:DDB1deltaBPB | | Descriptor: | Bromodomain-containing protein 4, DDB1- and CUL4-associated factor 16, DDB1deltaBPB, ... | | Authors: | Cowan, A.D, Sundaramoorthy, R, Nakasone, M.A, Ciulli, A. | | Deposit date: | 2023-04-25 | | Release date: | 2023-05-17 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.77 Å) | | Cite: | Targeted protein degradation via intramolecular bivalent glues.
Nature, 627, 2024
|
|
5AOV
 
 | |
8B1V
 
 | | Dihydroprecondylocarpine acetate synthase 2 from Tabernanthe iboga | | Descriptor: | Dihydroprecondylocarpine acetate synthase 2, ZINC ION, precondylocarpine acetate | | Authors: | Langley, C, Basquin, J, Caputi, L, O'Connor, S.E. | | Deposit date: | 2022-09-12 | | Release date: | 2022-10-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.882 Å) | | Cite: | Expansion of the Catalytic Repertoire of Alcohol Dehydrogenases in Plant Metabolism.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
8B25
 
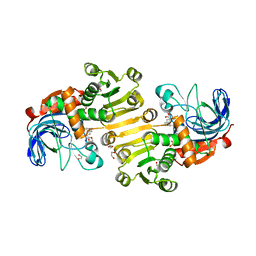 | | Dihydroprecondylocarpine acetate synthase 2 from Tabernanthe iboga - stemmadenine acetate bound structure | | Descriptor: | 1,2-ETHANEDIOL, Dihydroprecondylocarpine acetate synthase 2, SULFATE ION, ... | | Authors: | Langley, C, Basquin, J, Caputi, L, O'Connor, S.E. | | Deposit date: | 2022-09-13 | | Release date: | 2022-10-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Expansion of the Catalytic Repertoire of Alcohol Dehydrogenases in Plant Metabolism.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
8B26
 
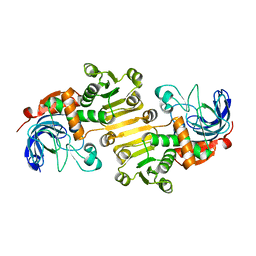 | | Dihydroprecondylocarpine acetate synthase 2 from Tabernanthe iboga | | Descriptor: | Dihydroprecondylocarpine acetate synthase 2, ZINC ION | | Authors: | Langley, C, Basquin, J, Caputi, L, O'Connor, S.E. | | Deposit date: | 2022-09-13 | | Release date: | 2022-10-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Expansion of the Catalytic Repertoire of Alcohol Dehydrogenases in Plant Metabolism.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
1RP3
 
 | |
6RJ8
 
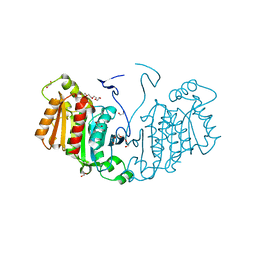 | | Structure of the alpha-beta hydrolase CorS from Tabernathe iboga | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, TETRAETHYLENE GLYCOL, ... | | Authors: | Farrow, S.C, Caputi, L, Kamileen, M.O, Bussey, K, Stevenson, C.E.M, Mundy, J, Lawson, D.M, O'Connor, S.E. | | Deposit date: | 2019-04-26 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Structural basis of cycloaddition in biosynthesis of iboga and aspidosperma alkaloids.
Nat.Chem.Biol., 16, 2020
|
|
1RMN
 
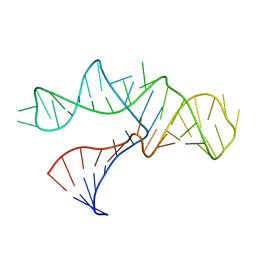 | | A THREE-DIMENSIONAL MODEL FOR THE HAMMERHEAD RIBOZYME BASED ON FLUORESCENCE MEASUREMENTS | | Descriptor: | RNA (49-MER) | | Authors: | Tuschl, T, Gohlke, C, Jovin, T.M, Westhof, E, Eckstein, F. | | Deposit date: | 1994-10-20 | | Release date: | 1995-01-26 | | Last modified: | 2024-02-14 | | Method: | FLUORESCENCE TRANSFER | | Cite: | A three-dimensional model for the hammerhead ribozyme based on fluorescence measurements.
Science, 266, 1994
|
|
7K14
 
 | | Ternary soak structure of alkanesulfonate monooxygenase MsuD from Pseudomonas fluorescens with FMN and methanesulfonate | | Descriptor: | Alkanesulfonate monooxygenase, CHLORIDE ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Liew, J.J.M, Dowling, D.P, El Saudi, I.M. | | Deposit date: | 2020-09-07 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structures of the alkanesulfonate monooxygenase MsuD provide insight into C-S bond cleavage, substrate scope, and an unexpected role for the tetramer.
J.Biol.Chem., 297, 2021
|
|
5DKW
 
 | |
6G2Q
 
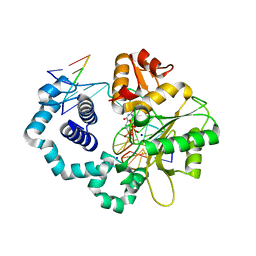 | | Ternary complex crystal structure of DNA polymerase Beta with a dideoxy terminated primer with CHCL (S-isomer), beta, gamma dTTP analogue | | Descriptor: | CHLORIDE ION, DNA polymerase beta, Downstream Primer Strand, ... | | Authors: | Batra, V.K, Wilson, S.H. | | Deposit date: | 2018-03-23 | | Release date: | 2018-07-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.148 Å) | | Cite: | Mapping Functional Substrate-Enzyme Interactions in the pol beta Active Site through Chemical Biology: Structural Responses to Acidity Modification of Incoming dNTPs.
Biochemistry, 57, 2018
|
|
