1EWI
 
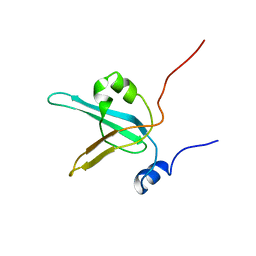 | | HUMAN REPLICATION PROTEIN A: GLOBAL FOLD OF THE N-TERMINAL RPA-70 DOMAIN REVEALS A BASIC CLEFT AND FLEXIBLE C-TERMINAL LINKER | | Descriptor: | REPLICATION PROTEIN A | | Authors: | Jacobs, D.M, Lipton, A.S, Isern, N.G, Daughdrill, G.W, Lowry, D.F, Gomes, X, Wold, M.S. | | Deposit date: | 2000-04-25 | | Release date: | 2000-05-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Human replication protein A: global fold of the N-terminal RPA-70 domain reveals a basic cleft and flexible C-terminal linker.
J.Biomol.NMR, 14, 1999
|
|
1NZM
 
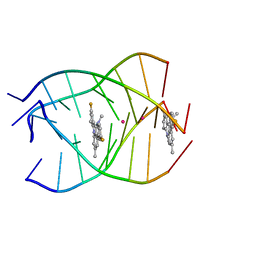 | | NMR structure of the parallel-stranded DNA quadruplex d(TTAGGGT)4 complexed with the telomerase inhibitor RHPS4 | | Descriptor: | 3,11-DIFLUORO-6,8,13-TRIMETHYL-8H-QUINO[4,3,2-KL]ACRIDIN-13-IUM, 5'-D(*TP*TP*AP*GP*GP*GP*T)-3', POTASSIUM ION | | Authors: | Gavathiotis, E, Heald, R.A, Stevens, M.F.G, Searle, M.S. | | Deposit date: | 2003-02-18 | | Release date: | 2003-11-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Drug Recognition and Stabilisation of the Parallel-stranded DNA Quadruplex d(TTAGGGT)4 Containing the Human Telomeric Repeat
J.Mol.Biol., 334, 2003
|
|
1EQ1
 
 | |
1E17
 
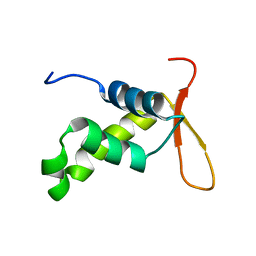 | | Solution structure of the DNA binding domain of the human Forkhead transcription factor AFX (FOXO4) | | Descriptor: | AFX | | Authors: | Weigelt, J, Climent, I, Dahlman-Wright, K, Wikstrom, M. | | Deposit date: | 2000-04-25 | | Release date: | 2000-08-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | 1H, 13C and 15N Resonance Assignments of the DNA Binding Domain of the Human Forkhead Transcription Factor Afx
J.Biomol.NMR, 17, 2000
|
|
5LW4
 
 | |
1EIT
 
 | | NMR STUDY OF MU-AGATOXIN | | Descriptor: | MU-AGATOXIN-I | | Authors: | Omecinsky, D.O, Reily, M.D. | | Deposit date: | 1995-12-14 | | Release date: | 1996-04-03 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure analysis of mu-agatoxins: further evidence for common motifs among neurotoxins with diverse ion channel specificities.
Biochemistry, 35, 1996
|
|
1EII
 
 | | NMR STRUCTURE OF HOLO CELLULAR RETINOL-BINDING PROTEIN II | | Descriptor: | CELLULAR RETINOL-BINDING PROTEIN II, RETINOL | | Authors: | Lu, J, Lin, C.L, Tang, C, Ponder, J.W, Kao, J.L, Cistola, D.P, Li, E. | | Deposit date: | 2000-02-25 | | Release date: | 2000-08-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Binding of retinol induces changes in rat cellular retinol-binding protein II conformation and backbone dynamics.
J.Mol.Biol., 300, 2000
|
|
4A1M
 
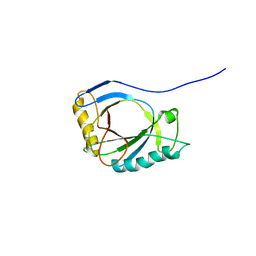 | | NMR Structure of protoporphyrin-IX bound murine p22HBP | | Descriptor: | HEME-BINDING PROTEIN 1 | | Authors: | Goodfellow, B.J, Dias, J.S, Macedo, A.L, Ferreira, G.C, Peterson, F.C, Volkman, B.F, Duarte, I.C.N. | | Deposit date: | 2011-09-15 | | Release date: | 2011-10-05 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The NMR Structure of Protoporphyrin-Ix Bound Murine P22Hbp
To be Published
|
|
1FH3
 
 | |
1M58
 
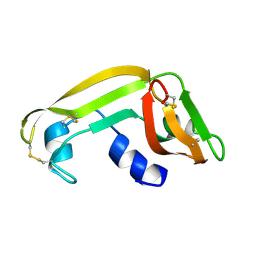 | | Solution Structure of Cytotoxic RC-RNase2 | | Descriptor: | RC-RNase2 ribonuclease | | Authors: | Hsu, C.-H, Liao, Y.-D, Wu, S.-H, Chen, C. | | Deposit date: | 2002-07-09 | | Release date: | 2003-01-09 | | Last modified: | 2022-12-21 | | Method: | SOLUTION NMR | | Cite: | 1H, 15N and 13C resonance assignments and secondary structure determination of the RC-RNase 2 from oocytes of bullfrog Rana catesbeiana.
J.Biomol.Nmr, 19, 2001
|
|
1G7D
 
 | | NMR STRUCTURE OF ERP29 C-DOMAIN | | Descriptor: | ENDOPLASMIC RETICULUM PROTEIN ERP29 | | Authors: | Liepinsh, E, Mkrtchian, S, Barishev, M, Sharipo, A, Ingelman-Sundberg, M, Otting, G. | | Deposit date: | 2000-11-10 | | Release date: | 2000-11-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Thioredoxin fold as homodimerization module in the putative chaperone ERp29: NMR structures of the domains and experimental model of the 51 kDa dimer.
Structure, 9, 2001
|
|
1G7E
 
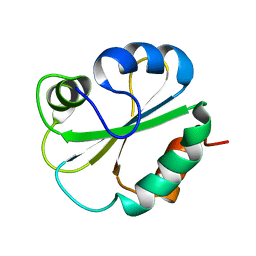 | | NMR STRUCTURE OF N-DOMAIN OF ERP29 PROTEIN | | Descriptor: | ENDOPLASMIC RETICULUM PROTEIN ERP29 | | Authors: | Liepinsh, E, Mkrtchian, S, Barishev, M, Sharipo, M, Ingelman-Sundberg, M, Otting, G. | | Deposit date: | 2000-11-10 | | Release date: | 2000-11-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Thioredoxin fold as homodimerization module in the putative chaperone ERp29: NMR structures of the domains and experimental model of the 51 kDa dimer.
Structure, 9, 2001
|
|
6FD7
 
 | |
1TIZ
 
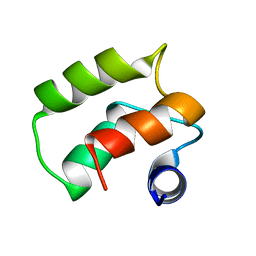 | | Solution Structure of a Calmodulin-Like Calcium-Binding Domain from Arabidopsis thaliana | | Descriptor: | calmodulin-related protein, putative | | Authors: | Song, J, Zhao, Q, Thao, S, Frederick, R.O, Markley, J.L, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2004-06-02 | | Release date: | 2004-08-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Letter to the Editor: Solution structure of a calmodulin-like calcium-binding domain from Arabidopsis thaliana
J.Biomol.NMR, 30, 2004
|
|
2M07
 
 | | NMR structure of OmpX in DPC micelles | | Descriptor: | Outer membrane protein X | | Authors: | Hagn, F.X, Etzkorn, M, Raschle, T, Wagner, G, Membrane Protein Structures by Solution NMR (MPSbyNMR) | | Deposit date: | 2012-10-21 | | Release date: | 2012-12-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Optimized phospholipid bilayer nanodiscs facilitate high-resolution structure determination of membrane proteins.
J.Am.Chem.Soc., 135, 2013
|
|
2M06
 
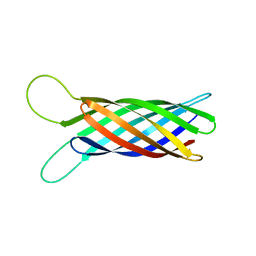 | | NMR structure of OmpX in phopspholipid nanodiscs | | Descriptor: | Outer membrane protein X | | Authors: | Hagn, F.X, Etzkorn, M, Raschle, T, Wagner, G, Membrane Protein Structures by Solution NMR (MPSbyNMR) | | Deposit date: | 2012-10-21 | | Release date: | 2012-12-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Optimized phospholipid bilayer nanodiscs facilitate high-resolution structure determination of membrane proteins.
J.Am.Chem.Soc., 135, 2013
|
|
1K19
 
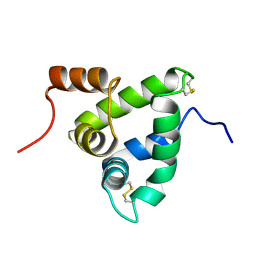 | | NMR Solution Structure of the Chemosensory Protein CSP2 from Moth Mamestra brassicae | | Descriptor: | Chemosensory Protein CSP2 | | Authors: | Mosbah, A, Campanacci, V, Lartigue, A, Tegoni, M, Cambillau, C, Darbon, H. | | Deposit date: | 2001-09-24 | | Release date: | 2002-12-04 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a chemosensory protein from the moth Mamestra brassicae
BIOCHEM.J., 369, 2003
|
|
1S4J
 
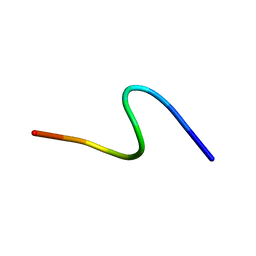 | | NMR structure of cross-reactive peptides from Homo sapiens | | Descriptor: | 60S acidic ribosomal protein P2 | | Authors: | Soares, M.R, Bisch, P.M, Campos de Carvalho, A.C, Valente, A.P, Almeida, F.C.L. | | Deposit date: | 2004-01-16 | | Release date: | 2004-03-16 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Correlation between conformation and antibody binding: NMR structure of cross-reactive peptides from T. cruzi, human and L. braziliensis
Febs Lett., 560, 2004
|
|
1S4H
 
 | | NMR structure of cross-reactive peptides from L. braziliensis | | Descriptor: | 60S acidic ribosomal protein P2 | | Authors: | Soares, M.R, Bisch, P.M, Campos de Carvalho, A.C, Valente, A.P, Almeida, F.C.L. | | Deposit date: | 2004-01-16 | | Release date: | 2004-03-16 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Correlation between conformation and antibody binding: NMR structure of cross-reactive peptides from T. cruzi, human and L. braziliensis
Febs Lett., 560, 2004
|
|
1B4M
 
 | | NMR STRUCTURE OF APO CELLULAR RETINOL-BINDING PROTEIN II, 24 STRUCTURES | | Descriptor: | CELLULAR RETINOL-BINDING PROTEIN II | | Authors: | Lu, J, Lin, C.-L, Tang, C, Ponder, J.W, Kao, J.L.F, Cistola, D.P, Li, E. | | Deposit date: | 1998-12-23 | | Release date: | 1999-04-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The structure and dynamics of rat apo-cellular retinol-binding protein II in solution: comparison with the X-ray structure.
J.Mol.Biol., 286, 1999
|
|
1CYZ
 
 | | NMR STRUCTURE OF THE GAACTGGTTC/TRI-IMIDAZOLE POLYAMIDE COMPLEX | | Descriptor: | (2-{[4-({4-[(4-FORMYLAMINO-1-METHYL-1H-IMIDAZOLE-2-CARBONYL)-AMINO]-1-METHYL-1H-IMIDAZOLE-2-CARBONYL}-AMINO)-1-METHYL-1 H-IMIDAZOLE-2-CARBONYL]-AMINO}-ETHYL)-DIMETHYL-AMMONIUM, 5'-D(*GP*AP*AP*CP*TP*GP*GP*TP*TP*C)-3' | | Authors: | Yang, X.-L, Hubbard IV, R.B, Lee, M, Tao, Z.-F, Sugiyama, H, Wang, A.H.-J. | | Deposit date: | 1999-08-31 | | Release date: | 1999-09-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Imidazole-imidazole pair as a minor groove recognition motif for T:G mismatched base pairs
Nucleic Acids Res., 27, 1999
|
|
1WWQ
 
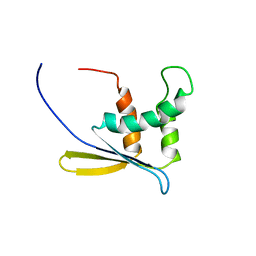 | | Solution Structure of Mouse ER | | Descriptor: | Enhancer of rudimentary homolog | | Authors: | Li, H, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-01-12 | | Release date: | 2006-01-03 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the mouse enhancer of rudimentary protein reveals a novel fold
J.Biomol.Nmr, 32, 2005
|
|
1E74
 
 | | NMR SOLUTION STRUCTURE OF ALPHA-CONOTOXIN IM1 POINT MUTATION VARIANT R11E | | Descriptor: | ALPHA-CONOTOXIN IM1(R11E) | | Authors: | Rogers, J.P, Luginbuhl, P, Pemberton, K, Harty, P, Wemmer, D.E, Stevens, R.C. | | Deposit date: | 2000-08-24 | | Release date: | 2000-12-27 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure-Activity Relationships in a Peptidic Alpha7 Nicotinic Acetylcholine Receptor Antagonist
J.Mol.Biol., 304, 2000
|
|
1E76
 
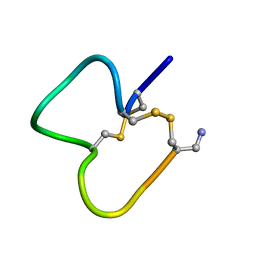 | | NMR SOLUTION STRUCTURE OF ALPHA-CONOTOXIN IM1 POINT MUTATION VARIANT D5N | | Descriptor: | ALPHA-CONOTOXIN IM1(D5N) | | Authors: | Rogers, J.P, Luginbuhl, P, Pemberton, K, Harty, P, Wemmer, D.E, Stevens, R.C. | | Deposit date: | 2000-08-24 | | Release date: | 2000-12-27 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure-Activity Relationships in a Peptidic Alpha7 Nicotinic Acetylcholine Receptor Antagonist
J.Mol.Biol., 304, 2000
|
|
1E75
 
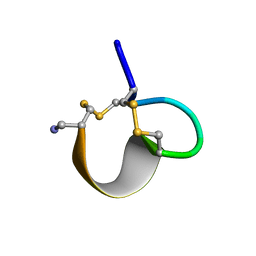 | | NMR SOLUTION STRUCTURE OF ALPHA-CONOTOXIN IM1 POINT MUTATION VARIANT R7L | | Descriptor: | ALPHA-CONOTOXIN IM1(R7L) | | Authors: | Rogers, J.P, Luginbuhl, P, Pemberton, K, Harty, P, Wemmer, D.E, Stevens, R.C. | | Deposit date: | 2000-08-24 | | Release date: | 2000-12-27 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure-Activity Relationships in a Peptidic Alpha7 Nicotinic Acetylcholine Receptor Antagonist
J.Mol.Biol., 304, 2000
|
|
