2F3E
 
 | | Crystal Structure of the Bace complex with AXQ093, a macrocyclic inhibitor | | Descriptor: | Beta-secretase 1, {(E)-(3R,6S,9R)-3-[(1S,3R)-3-((S)-1 -BUTYLCARBAMOYL-2-METHYL-PROPYLCARB AMOYL)-1-HYDROXY-BUTYL]-6-METHYL-5, 8-DIOXO-1,11-DITHIA-4,7-DIAZA-CYCLO PENTADEC-13-EN-9-YL}-CARBAMIC ACID TERT-BUTYL ESTER | | Authors: | Rondeau, J.-M. | | Deposit date: | 2005-11-21 | | Release date: | 2006-09-05 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structure-based design and synthesis of macroheterocyclic peptidomimetic inhibitors of the aspartic protease beta-site amyloid precursor protein cleaving enzyme (BACE).
J.Med.Chem., 49, 2006
|
|
2F3Y
 
 | | Calmodulin/IQ domain complex | | Descriptor: | CALCIUM ION, Calmodulin, MAGNESIUM ION, ... | | Authors: | Fallon, J.L, Quiocho, F.A. | | Deposit date: | 2005-11-22 | | Release date: | 2005-12-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of Calmodulin Bound to the Hydrophobic IQ Domain of the Cardiac Ca(v)1.2 Calcium Channel.
Structure, 13, 2005
|
|
2F7A
 
 | | BenM effector binding domain with its effector, cis,cis-muconate | | Descriptor: | (2Z,4Z)-HEXA-2,4-DIENEDIOIC ACID, ACETATE ION, BENZOIC ACID, ... | | Authors: | Clark, T, Haddad, S, Ezezika, O, Neidle, E, Momany, C. | | Deposit date: | 2005-11-30 | | Release date: | 2006-10-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Distinct Effector-binding Sites Enable Synergistic Transcriptional Activation by BenM, a LysR-type Regulator.
J.Mol.Biol., 367, 2007
|
|
2I55
 
 | |
2HRW
 
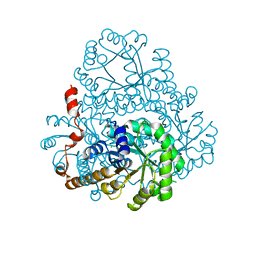 | | Crystal Structure of Phosphonopyruvate Hydrolase | | Descriptor: | CHLORIDE ION, Phosphonopyruvate hydrolase, SODIUM ION | | Authors: | Chen, C.C.H, Herzberg, O. | | Deposit date: | 2006-07-20 | | Release date: | 2006-10-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and Kinetics of Phosphonopyruvate Hydrolase from Voriovorax sp. Pal2: New Insight into the Divergence of Catalysis within the PEP Mutase/Isocitrate Lyase Superfamily
Biochemistry, 45, 2006
|
|
2HRT
 
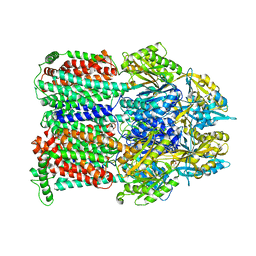 | | Asymmetric structure of trimeric AcrB from Escherichia coli | | Descriptor: | Acriflavine resistance protein B, CITRATE ANION | | Authors: | Seeger, M.A, Schiefner, A, Eicher, T, Verrey, F, Diederichs, K, Pos, K.M. | | Deposit date: | 2006-07-20 | | Release date: | 2006-09-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Asymmetry of AcrB Trimer Suggests a Peristaltic Pump Mechanism.
Science, 313, 2006
|
|
2HUW
 
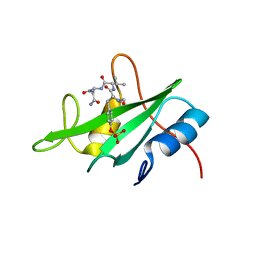 | |
2I6K
 
 | | Crystal structure of human type I IPP isomerase complexed with a substrate analog | | Descriptor: | ACETIC ACID, AMINOETHANOLPYROPHOSPHATE, Isopentenyl-diphosphate delta-isomerase 1, ... | | Authors: | Zhang, C, Wei, Z, Gong, W. | | Deposit date: | 2006-08-29 | | Release date: | 2007-06-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of human IPP isomerase: new insights into the catalytic mechanism
J.Mol.Biol., 366, 2007
|
|
2I74
 
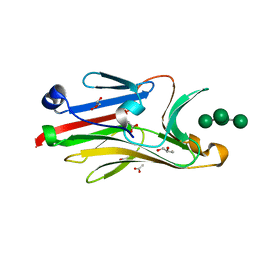 | | Crystal structure of mouse Peptide N-Glycanase C-terminal domain in complex with mannopentaose | | Descriptor: | ACETATE ION, GLYCEROL, PNGase, ... | | Authors: | Zhou, X, Zhao, G, Wang, L, Li, G, Lennarz, W.J, Schindelin, H. | | Deposit date: | 2006-08-30 | | Release date: | 2006-10-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural and biochemical studies of the C-terminal domain of mouse peptide-N-glycanase identify it as a mannose-binding module.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2I7F
 
 | | Sphingomonas yanoikuyae B1 ferredoxin | | Descriptor: | CITRIC ACID, FE2/S2 (INORGANIC) CLUSTER, Ferredoxin component of dioxygenase, ... | | Authors: | Ramaswamy, S, Brown, E.N. | | Deposit date: | 2006-08-30 | | Release date: | 2007-03-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural investigations of the ferredoxin and terminal oxygenase components of the biphenyl 2,3-dioxygenase from Sphingobium yanoikuyae B1.
Bmc Struct.Biol., 7, 2007
|
|
2G6W
 
 | |
2G78
 
 | |
2GAK
 
 | |
2GB4
 
 | |
2GBT
 
 | | C6A/C111A CuZn Superoxide dismutase | | Descriptor: | COPPER (I) ION, Superoxide dismutase [Cu-Zn], ZINC ION | | Authors: | Hornberg, A, Logan, D.T, Marklund, S.L, Oliveberg, M. | | Deposit date: | 2006-03-11 | | Release date: | 2007-01-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Coupling between Disulphide Status, Metallation and Dimer Interface Strength in Cu/Zn Superoxide Dismutase
J.Mol.Biol., 365, 2007
|
|
2G7B
 
 | |
2GFH
 
 | |
2GBI
 
 | | rat DPP-IV with xanthine inhibitor 4 | | Descriptor: | 2-({8-[(3R)-3-AMINOPIPERIDIN-1-YL]-1,3-DIMETHYL-2,6-DIOXO-1,2,3,6-TETRAHYDRO-7H-PURIN-7-YL}METHYL)BENZONITRILE, Dipeptidyl peptidase 4 | | Authors: | Longenecker, K.L, Jakob, C.G, Fry, E.H, Wilk, S. | | Deposit date: | 2006-03-10 | | Release date: | 2006-07-04 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal Structures of DPP-IV (CD26) from Rat Kidney Exhibit Flexible Accommodation of Peptidase-Selective Inhibitors.
Biochemistry, 45, 2006
|
|
2GHS
 
 | |
2G30
 
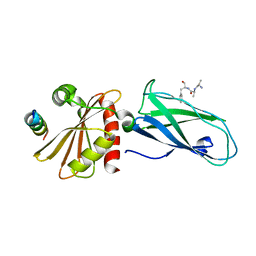 | | beta appendage of AP2 complexed with ARH peptide | | Descriptor: | 16-mer peptide from Low density lipoprotein receptor adapter protein 1, AP-2 complex subunit beta-1, peptide sequence AAF | | Authors: | Edeling, M.A, Collins, B.M, Traub, L.M, Owen, D.J. | | Deposit date: | 2006-02-17 | | Release date: | 2006-03-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular Switches Involving the AP-2 beta2 Appendage Regulate Endocytic Cargo Selection and Clathrin Coat Assembly
Dev.Cell, 10, 2006
|
|
2G0C
 
 | |
2G50
 
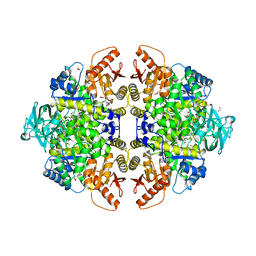 | | The location of the allosteric amino acid binding site of muscle pyruvate kinase. | | Descriptor: | 1,2-ETHANEDIOL, 2-{2-[2-2-(METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, ALANINE, ... | | Authors: | Holyoak, T, Williams, R, Fenton, A.W. | | Deposit date: | 2006-02-22 | | Release date: | 2006-05-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Differentiating a Ligand's Chemical Requirements for Allosteric Interactions from Those for Protein Binding. Phenylalanine Inhibition of Pyruvate Kinase.
Biochemistry, 45, 2006
|
|
2GA1
 
 | |
2FEU
 
 | | P450CAM from Pseudomonas putida reconstituted with manganic protoporphyrin IX | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CAMPHOR, Cytochrome P450-cam, ... | | Authors: | von Koenig, K, Makris, T.M, Sligar, S.G, Schlichting, I. | | Deposit date: | 2005-12-16 | | Release date: | 2006-03-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The status of high-valent metal oxo complexes in the P450 cytochromes.
J.Inorg.Biochem., 100, 2006
|
|
2FJA
 
 | | adenosine 5'-phosphosulfate reductase in complex with substrate | | Descriptor: | ADENOSINE-5'-PHOSPHOSULFATE, FLAVIN-ADENINE DINUCLEOTIDE, IRON/SULFUR CLUSTER, ... | | Authors: | Schiffer, A, Fritz, G, Kroneck, P.M, Ermler, U. | | Deposit date: | 2006-01-02 | | Release date: | 2006-03-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Reaction mechanism of the iron-sulfur flavoenzyme adenosine-5'-phosphosulfate reductase based on the structural characterization of different enzymatic states
Biochemistry, 45, 2006
|
|
