5KLO
 
 | | Crystal structure of thioacyl intermediate in 2-aminomuconate 6-semialdehyde dehydrogenase N169A | | Descriptor: | (2Z,4E)-2-hydroxy-6-oxohexa-2,4-dienoic acid, 2-aminomuconate 6-semialdehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Yang, Y, Davis, I, Ha, U, Wang, Y, Shin, I, Liu, A. | | Deposit date: | 2016-06-24 | | Release date: | 2016-11-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | A Pitcher-and-Catcher Mechanism Drives Endogenous Substrate Isomerization by a Dehydrogenase in Kynurenine Metabolism.
J.Biol.Chem., 291, 2016
|
|
4ZIT
 
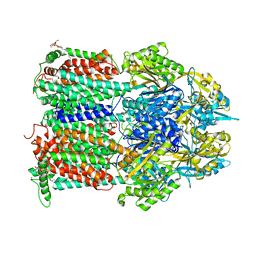 | | Crystal structure of AcrB in P21 space group | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Multidrug efflux pump subunit AcrB, NICKEL (II) ION | | Authors: | Ababou, A, Koronakis, V. | | Deposit date: | 2015-04-28 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.296 Å) | | Cite: | Structures of Gate Loop Variants of the AcrB Drug Efflux Pump Bound by Erythromycin Substrate.
Plos One, 11, 2016
|
|
3LC2
 
 | | Crystal Structure of Thioacyl-Glyceraldehyde-3-phosphate dehydrogenase 1(GAPDH 1) from methicillin resistant Staphylococcus aureus MRSA252 | | Descriptor: | CHLORIDE ION, GLYCERALDEHYDE-3-PHOSPHATE, GLYCEROL, ... | | Authors: | Mukherjee, S, Dutta, D, Saha, B, Das, A.K. | | Deposit date: | 2010-01-09 | | Release date: | 2010-08-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of glyceraldehyde-3-phosphate dehydrogenase 1 from methicillin-resistant Staphylococcus aureus MRSA252 provides novel insights into substrate binding and catalytic mechanism.
J.Mol.Biol., 401, 2010
|
|
5L1H
 
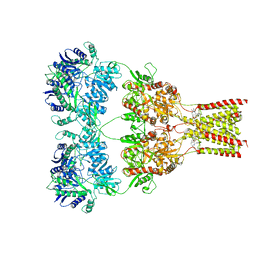 | | AMPA subtype ionotropic glutamate receptor GluA2 in complex with noncompetitive inhibitor GYKI53655 | | Descriptor: | (8R)-5-(4-aminophenyl)-N,8-dimethyl-8,9-dihydro-2H,7H-[1,3]dioxolo[4,5-h][2,3]benzodiazepine-7-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor 2 | | Authors: | Yelshanskaya, M.V, Singh, A.K, Sampson, J.M, Sobolevsky, A.I. | | Deposit date: | 2016-07-29 | | Release date: | 2016-10-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.801 Å) | | Cite: | Structural Bases of Noncompetitive Inhibition of AMPA-Subtype Ionotropic Glutamate Receptors by Antiepileptic Drugs.
Neuron, 91, 2016
|
|
7T8J
 
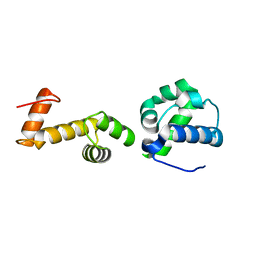 | | The ubiquitin-associated domain of human thirty-eight negative kinase-1 flexibly fused to the 1TEL crystallization chaperone via a GSGG linker | | Descriptor: | CHLORIDE ION, Transcription factor ETV6,Non-receptor tyrosine-protein kinase TNK1 | | Authors: | Soleimani, S, Pedroza Romo, M.J, Smith, T, Brown, S, Doukov, T, Moody, J.D. | | Deposit date: | 2021-12-16 | | Release date: | 2022-12-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Fusion crystallization reveals the behavior of both the 1TEL crystallization chaperone and the TNK1 UBA domain.
Structure, 31, 2023
|
|
3ERP
 
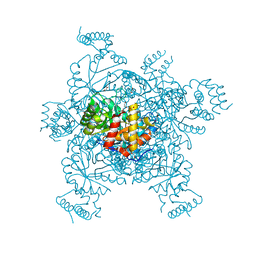 | | Structure of IDP01002, a putative oxidoreductase from and essential gene of Salmonella typhimurium | | Descriptor: | 1,2-ETHANEDIOL, CACODYLATE ION, CHLORIDE ION, ... | | Authors: | Singer, A.U, Minasov, G, Evdokimova, E, Brunzelle, J.S, Kudritska, M, Edwards, A.M, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2008-10-02 | | Release date: | 2008-11-04 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural and biochemical studies of novel aldo-keto reductases for the biocatalytic conversion of 3-hydroxybutanal to 1,3-butanediol.
Appl.Environ.Microbiol., 2017
|
|
7T5C
 
 | |
7T5E
 
 | |
7FAW
 
 | | Structure of LW domain from Yeast | | Descriptor: | Transcription elongation factor S-II | | Authors: | Liao, S, Gao, J, Tu, X. | | Deposit date: | 2021-07-07 | | Release date: | 2022-07-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.438 Å) | | Cite: | Structural basis for evolutionarily conserved interactions between TFIIS and Paf1C.
Int.J.Biol.Macromol., 253, 2023
|
|
3LB8
 
 | |
7T5D
 
 | |
5KXT
 
 | | Hen Egg White Lysozyme at 278K, Data set 5 | | Descriptor: | Lysozyme C, SODIUM ION | | Authors: | Russi, S, Gonzalez, A, Kenner, L.R, Keedy, D.A, Fraser, J.S, van den Bedem, H. | | Deposit date: | 2016-07-20 | | Release date: | 2016-09-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Conformational variation of proteins at room temperature is not dominated by radiation damage.
J Synchrotron Radiat, 24, 2017
|
|
1WID
 
 | | Solution Structure of the B3 DNA-Binding Domain of RAV1 | | Descriptor: | DNA-binding protein RAV1 | | Authors: | Yamasaki, K, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-28 | | Release date: | 2004-11-28 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the B3 DNA Binding Domain of the Arabidopsis Cold-Responsive Transcription Factor RAV1
Plant Cell, 16, 2004
|
|
7TDY
 
 | | The ubiquitin-associated domain of human thirty-eight negative kinase 1, flexibly fused to the 1TEL crystallization chaperone via a 2-glycine linker and crystallized at low protein concentration | | Descriptor: | FORMIC ACID, Transcription factor ETV6,Non-receptor tyrosine-protein kinase TNK1 | | Authors: | Nawarathnage, S, Bunn, D.R, Stewart, C, Doukev, T, Moody, J.D. | | Deposit date: | 2022-01-03 | | Release date: | 2023-01-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Fusion crystallization reveals the behavior of both the 1TEL crystallization chaperone and the TNK1 UBA domain.
Structure, 31, 2023
|
|
5YIL
 
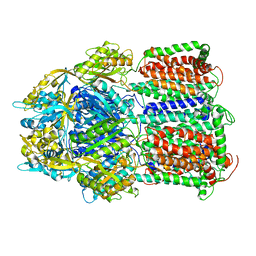 | | Hoisting-loop in bacterial multidrug exporter AcrB is a highly flexible hinge that enables the large motion of the subdomains | | Descriptor: | Multidrug efflux pump subunit AcrB | | Authors: | Zwama, M, Sakurai, K, Hayashi, K, Nakashima, R, Kitagawa, K, Nishino, K, Yamaguchi, A. | | Deposit date: | 2017-10-05 | | Release date: | 2017-11-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Hoisting-Loop in Bacterial Multidrug Exporter AcrB Is a Highly Flexible Hinge That Enables the Large Motion of the Subdomains.
Front Microbiol, 8, 2017
|
|
7TCY
 
 | | The ubiquitin-associated domain of human thirty-eight negative kinase I | | Descriptor: | CHLORIDE ION, FORMIC ACID, MAGNESIUM ION, ... | | Authors: | Nawarathnage, S, Bunn, R.D, Stewart, C, Doukov, T, Moody, J.D. | | Deposit date: | 2021-12-29 | | Release date: | 2023-01-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Fusion crystallization reveals the behavior of both the 1TEL crystallization chaperone and the TNK1 UBA domain.
Structure, 31, 2023
|
|
7F72
 
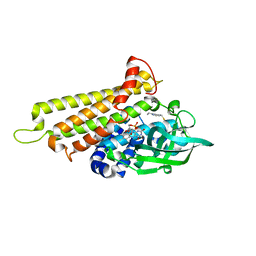 | | Rv3094c in complex with FAD and ETH. | | Descriptor: | 2-ethylpyridine-4-carboximidothioic acid, FLAVIN MONONUCLEOTIDE, Rv3094c | | Authors: | Wang, Z.X, Ouyang, S.Y. | | Deposit date: | 2021-06-27 | | Release date: | 2022-07-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Omics analysis of Mycobacterium tuberculosis isolates uncovers Rv3094c, an ethionamide metabolism-associated gene.
Commun Biol, 6, 2023
|
|
5L1B
 
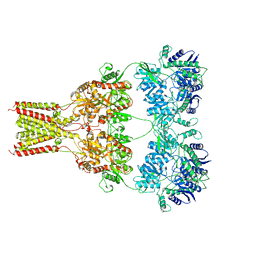 | | AMPA subtype ionotropic glutamate receptor GluA2 in Apo state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor 2,Glutamate receptor 2 | | Authors: | Yelshanskaya, M.V, Singh, A.K, Sampson, J.M, Sobolevsky, A.I. | | Deposit date: | 2016-07-28 | | Release date: | 2016-10-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structural Bases of Noncompetitive Inhibition of AMPA-Subtype Ionotropic Glutamate Receptors by Antiepileptic Drugs.
Neuron, 91, 2016
|
|
7TC7
 
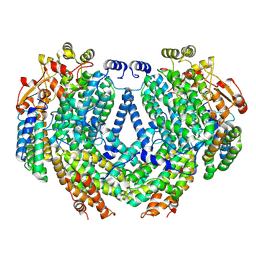 | | Cryo-EM structure of methane monooxygenase hydroxylase (by quantifoil) | | Descriptor: | FE (III) ION, Methane monooxygenase component A alpha chain, Methane monooxygenase component A beta chain, ... | | Authors: | Cho, U.S, Kim, B.C. | | Deposit date: | 2021-12-23 | | Release date: | 2023-01-25 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Batch Production of High-Quality Graphene Grids for Cryo-EM: Cryo-EM Structure of Methylococcus capsulatus Soluble Methane Monooxygenase Hydroxylase.
Acs Nano, 17, 2023
|
|
3LHL
 
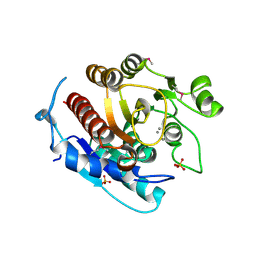 | | Crystal structure of a putative agmatinase from Clostridium difficile | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, MANGANESE (II) ION, PHOSPHATE ION, ... | | Authors: | Palani, K, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-01-22 | | Release date: | 2010-02-23 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a putative agmatinase from Clostridium difficile
To be Published
|
|
5L3H
 
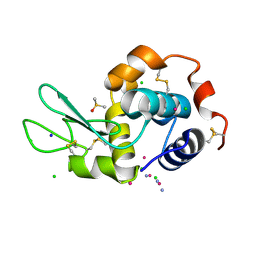 | |
7TC8
 
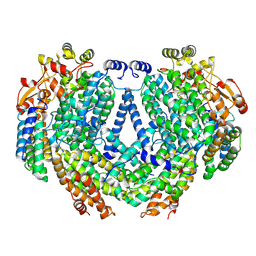 | | Cryo-EM structure of methane monooxygenase hydroxylase (by graphene) | | Descriptor: | FE (III) ION, Methane monooxygenase component A alpha chain, Methane monooxygenase component A beta chain, ... | | Authors: | Cho, U.S, Kim, B.C. | | Deposit date: | 2021-12-23 | | Release date: | 2023-01-25 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Batch Production of High-Quality Graphene Grids for Cryo-EM: Cryo-EM Structure of Methylococcus capsulatus Soluble Methane Monooxygenase Hydroxylase.
Acs Nano, 17, 2023
|
|
5L57
 
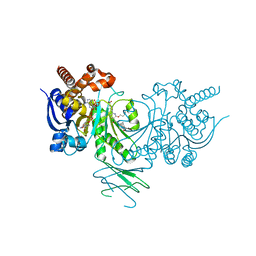 | | Crystal structure of Iso-citrate Dehydrogenase R132H in complex with a novel inhibitor (compound 13a) | | Descriptor: | (1~{R},5~{S})-3-[6-(3-methylbutoxy)-5-[[(1~{R},3~{S})-5-oxidanyl-2-adamantyl]carbamoyl]pyridin-2-yl]-3-azabicyclo[3.1.0]hexane-6-carboxylic acid, Isocitrate dehydrogenase [NADP] cytoplasmic, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Levy, C. | | Deposit date: | 2016-05-28 | | Release date: | 2016-12-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.695 Å) | | Cite: | Discovery and Optimization of Allosteric Inhibitors of Mutant Isocitrate Dehydrogenase 1 (R132H IDH1) Displaying Activity in Human Acute Myeloid Leukemia Cells.
J.Med.Chem., 59, 2016
|
|
3LKX
 
 | | Human nac dimerization domain | | Descriptor: | Nascent polypeptide-associated complex subunit alpha, Transcription factor BTF3 | | Authors: | Liu, Y, Hu, Y, Li, X, Niu, L, Teng, M. | | Deposit date: | 2010-01-28 | | Release date: | 2010-03-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of the human nascent polypeptide-associated complex domain reveals a nucleic acid-binding region on the NACA subunit
Biochemistry, 49, 2010
|
|
5KLM
 
 | | Crystal structure of 2-hydroxymuconate-6-semialdehyde derived intermediate in NAD(+)-bound 2-aminomuconate 6-semialdehyde dehydrogenase N169D | | Descriptor: | 2-aminomuconate 6-semialdehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION | | Authors: | Yang, Y, Davis, I, Ha, U, Wang, Y, Shin, I, Liu, A. | | Deposit date: | 2016-06-24 | | Release date: | 2016-11-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | A Pitcher-and-Catcher Mechanism Drives Endogenous Substrate Isomerization by a Dehydrogenase in Kynurenine Metabolism.
J.Biol.Chem., 291, 2016
|
|
