3K45
 
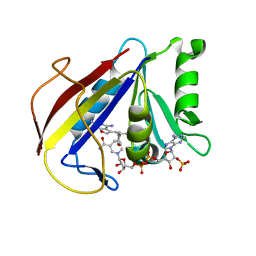 | | Alternate Binding Modes Observed for the E- and Z-isomers of 2,4-Diaminofuro[2,3d]pyrimidines as Ternary Complexes with NADPH and Mouse Dihydrofolate Reductase | | Descriptor: | 5-[(1Z)-2-(2-methoxyphenyl)prop-1-en-1-yl]furo[2,3-d]pyrimidine-2,4-diamine, Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cody, V. | | Deposit date: | 2009-10-05 | | Release date: | 2009-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Design, synthesis, and X-ray crystal structures of 2,4-diaminofuro[2,3-d]pyrimidines as multireceptor tyrosine kinase and dihydrofolate reductase inhibitors.
Bioorg.Med.Chem., 17, 2009
|
|
3K47
 
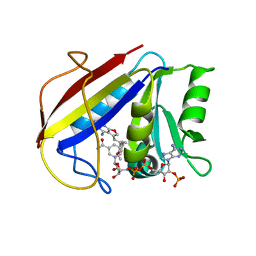 | | Alternate Binding Modes Observed for the E- and Z-Isomers of 2,4-Diaminofuro[2,3-d]pyrimidines as Ternary Complexes with NADPH and Mouse Dihydrofolate Reductase | | Descriptor: | 5-[(1E)-2-(2-methoxyphenyl)prop-1-en-1-yl]furo[2,3-d]pyrimidine-2,4-diamine, Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cody, V, Pace, J, Queener, S.F, Gangjee, A. | | Deposit date: | 2009-10-05 | | Release date: | 2009-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Design, synthesis, and X-ray crystal structures of 2,4-diaminofuro[2,3-d]pyrimidines as multireceptor tyrosine kinase and dihydrofolate reductase inhibitors.
Bioorg.Med.Chem., 17, 2009
|
|
1VD8
 
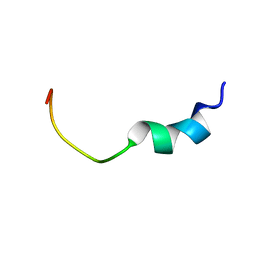 | | Solution structure of FMBP-1 tandem repeat 2 | | Descriptor: | fibroin-modulator-binding-protein-1 | | Authors: | Yamaki, T, Kawaguchi, K, Aizawa, T, Takiya, S, Demura, M, Nitta, K. | | Deposit date: | 2004-03-20 | | Release date: | 2005-03-29 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of FMBP-1 tandem repeat 2
To be Published
|
|
1VD7
 
 | | Solution structure of FMBP-1 tandem repeat 1 | | Descriptor: | Fibroin-modulator-binding-protein-1 | | Authors: | Kawaguchi, K, Yamaki, T, Aizawa, T, Takiya, S, Demura, M, Nitta, K. | | Deposit date: | 2004-03-20 | | Release date: | 2005-03-29 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of FMBP-1 tandem repeat 1
To be Published
|
|
1VLC
 
 | |
5W4X
 
 | | Truncated hUGDH | | Descriptor: | 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, UDP-glucose 6-dehydrogenase | | Authors: | Sennett, N.C, Custer, G.S, Wood, Z.A. | | Deposit date: | 2017-06-13 | | Release date: | 2017-07-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The entropic force generated by intrinsically disordered segments tunes protein function.
Nature, 563, 2018
|
|
3JWC
 
 | | Crystal structure of Bacillus anthracis (Y102F) dihydrofolate reductase complexed with NADPH and 2,4-diamino-5-(3-(3,4,5-trimethoxyphenyl)prop-1-ynyl)-6-ethylpyrimidine (UCP120A) | | Descriptor: | 6-ethyl-5-[3-(3,4,5-trimethoxyphenyl)prop-1-yn-1-yl]pyrimidine-2,4-diamine, Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Anderson, A.C, Beierlein, J.M, Karri, N.G. | | Deposit date: | 2009-09-18 | | Release date: | 2010-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Targeted Mutations in Bacillus anthracis Dihydrofolate Reductase Condense Complex Structure-Activity Relationships
J.Med.Chem., 2010
|
|
3JWK
 
 | | Crystal structure of Bacillus anthracis (Y102F) dihydrofolate reductase complexed with NADPH and (S)-2,4-diamino-5-(3-methoxy-3-(3,4,5-trimethoxyphenyl)prop-1-ynyl)-6-methylpyrimidine (UCP114A) | | Descriptor: | 5-[(3S)-3-methoxy-3-(3,4,5-trimethoxyphenyl)prop-1-yn-1-yl]-6-methylpyrimidine-2,4-diamine, Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Anderson, A.C, Beierlein, J.M, Karri, N.G. | | Deposit date: | 2009-09-18 | | Release date: | 2010-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Targeted Mutations in Bacillus anthracis Dihydrofolate Reductase Condense Complex Structure-Activity Relationships
J.Med.Chem., 2010
|
|
5WHL
 
 | |
1W27
 
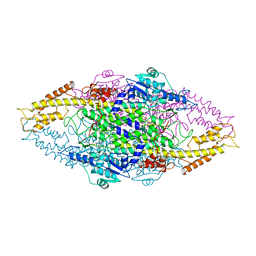 | | Phenylalanine ammonia-lyase (PAL) from Petroselinum crispum | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, PHENYLALANINE AMMONIA-LYASE 1 | | Authors: | Ritter, H, Schulz, G.E. | | Deposit date: | 2004-06-29 | | Release date: | 2004-11-25 | | Last modified: | 2025-10-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis for the Entrance Into the Phenylpropanoid Metabolism Catalyzed by Phenylalanine Ammonia-Lyase
Plant Cell, 16, 2004
|
|
5WEH
 
 | | Cytochrome c oxidase from Rhodobacter sphaeroides in the reduced state | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, Aa3-type cytochrome c oxidase subunit IV, CALCIUM ION, ... | | Authors: | Liu, J, Ferguson-Miller, F, Ling, Q, Hiser, C. | | Deposit date: | 2017-07-10 | | Release date: | 2017-09-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Role of conformational change and K-path ligands in controlling cytochrome c oxidase activity.
Biochem. Soc. Trans., 45, 2017
|
|
1W3X
 
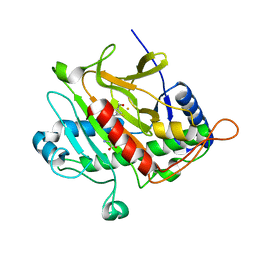 | | Isopenicillin N synthase d-(L-a-aminoadipoyl)-(3R)-methyl-L-cysteine D-a-hydroxyisovaleryl ester complex (Oxygen exposed 5 minutes 20 bar) | | Descriptor: | FE (II) ION, ISOPENICILLIN N SYNTHETASE, N~6~-[(1R)-1-({[(1R,2R)-1-CARBOXY-3-HYDROXY-2-METHYLPROPYL]OXY}CARBONYL)-2-MERCAPTOPROP-2-EN-1-YL]-6-OXO-L-LYSINE | | Authors: | Daruzzaman, A, Clifton, I.J, Rutledge, P.J. | | Deposit date: | 2004-07-20 | | Release date: | 2005-12-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Unexpected Oxidation of a Depsipeptide Substrate Analogue in Crystalline Isopenicillin N Synthase.
Chembiochem, 7, 2006
|
|
3JQ9
 
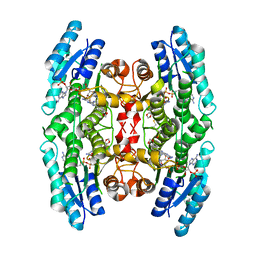 | | Crystal structure of pteridine reductase 1 (PTR1) from Trypanosoma brucei in ternary complex with cofactor (NADP+) and inhibitor 2-amino-6-(1,3-benzodioxol-5-yl)-4-oxo-4,7-dihydro-3H-pyrrolo[2,3-d]pyrimidine-5-carbonitrile (AX1) | | Descriptor: | 2-amino-6-(1,3-benzodioxol-5-yl)-4-oxo-4,7-dihydro-3H-pyrrolo[2,3-d]pyrimidine-5-carbonitrile, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Pteridine reductase 1 | | Authors: | Tulloch, L.B, Hunter, W.N. | | Deposit date: | 2009-09-06 | | Release date: | 2009-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-based design of pteridine reductase inhibitors targeting african sleeping sickness and the leishmaniases.
J.Med.Chem., 53, 2010
|
|
5W8K
 
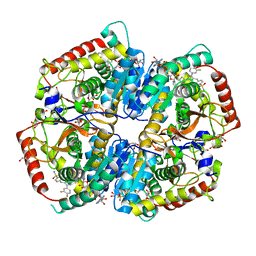 | | Crystal Structure of Lactate Dehydrogenase A in complex with inhibitor compound 29 and NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 2-{3-(3,4-difluorophenyl)-5-hydroxy-4-[(4-sulfamoylphenyl)methyl]-1H-pyrazol-1-yl}-1,3-thiazole-4-carboxylic acid, GLYCEROL, ... | | Authors: | Lukacs, C.M, Dranow, D.M. | | Deposit date: | 2017-06-21 | | Release date: | 2018-01-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery and Optimization of Potent, Cell-Active Pyrazole-Based Inhibitors of Lactate Dehydrogenase (LDH).
J. Med. Chem., 60, 2017
|
|
5LYK
 
 | | CRYSTAL STRUCTURE OF INTRACELLULAR B30.2 DOMAIN OF BTN3A1 BOUND TO CITRATE | | Descriptor: | 1,2-ETHANEDIOL, Butyrophilin subfamily 3 member A1, CITRATE ANION | | Authors: | Mohammed, F, Baker, A.T, Salim, M, Willcox, B.E. | | Deposit date: | 2016-09-28 | | Release date: | 2017-09-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | BTN3A1 Discriminates gamma delta T Cell Phosphoantigens from Nonantigenic Small Molecules via a Conformational Sensor in Its B30.2 Domain.
ACS Chem. Biol., 12, 2017
|
|
5W21
 
 | | Crystal Structure of a 1:1:1 FGF23-FGFR1c-aKlotho Ternary Complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fibroblast growth factor 23, Fibroblast growth factor receptor 1, ... | | Authors: | Mohammadi, M. | | Deposit date: | 2017-06-05 | | Release date: | 2018-01-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | alpha-Klotho is a non-enzymatic molecular scaffold for FGF23 hormone signalling.
Nature, 553, 2018
|
|
5LXE
 
 | | F420-dependent glucose-6-phosphate dehydrogenase from Rhodococcus jostii RHA1 | | Descriptor: | F420-dependent glucose-6-phosphate dehydrogenase 1, GLYCEROL, SULFATE ION | | Authors: | Nguyen, Q.-T, Trinco, G, Binda, C, Mattevi, A, Fraaije, M.W. | | Deposit date: | 2016-09-20 | | Release date: | 2016-12-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Discovery and characterization of an F420-dependent glucose-6-phosphate dehydrogenase (Rh-FGD1) from Rhodococcus jostii RHA1.
Appl. Microbiol. Biotechnol., 101, 2017
|
|
5LOQ
 
 | | Structure of coproheme bound HemQ from Listeria monocytogenes | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,3,5,8-TETRAMETHYL-PORPHINE-2,4,6,7-TETRAPROPIONIC ACID FERROUS COMPLEX, Putative heme-dependent peroxidase lmo2113, ... | | Authors: | Puehringer, D, Mlynek, G, Hofbauer, S, Djinovic-Carugo, K, Obinger, C. | | Deposit date: | 2016-08-09 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Hydrogen peroxide-mediated conversion of coproheme to heme b by HemQ-lessons from the first crystal structure and kinetic studies.
FEBS J., 283, 2016
|
|
5W97
 
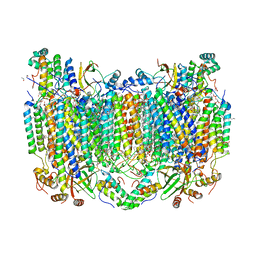 | | Crystal Structure of CO-bound Cytochrome c Oxidase determined by Serial Femtosecond X-Ray Crystallography at Room Temperature | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Rousseau, D.L, Yeh, S.-R, Ishigami, I, Zatsepin, N.A, Grant, T.D, Fromme, P, Fromme, R. | | Deposit date: | 2017-06-22 | | Release date: | 2017-08-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of CO-bound cytochrome c oxidase determined by serial femtosecond X-ray crystallography at room temperature.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5M0K
 
 | |
5M12
 
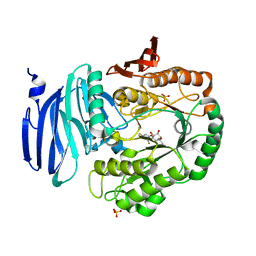 | | Structure of GH36 alpha-galactosidase from Thermotoga maritima in complex with intact cyclopropyl-carbasugar. | | Descriptor: | (1~{R},2~{S},3~{S},4~{R},5~{S},6~{S})-5-[3,5-bis(fluoranyl)phenoxy]-1-(hydroxymethyl)bicyclo[4.1.0]heptane-2,3,4-triol, Alpha-galactosidase, MAGNESIUM ION, ... | | Authors: | Pengelly, R, Gloster, T. | | Deposit date: | 2016-10-07 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Structural Snapshots for Mechanism-Based Inactivation of a Glycoside Hydrolase by Cyclopropyl Carbasugars.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
7L0M
 
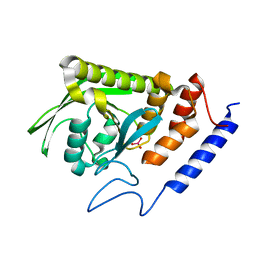 | | Vanadate-bound YopH G352T | | Descriptor: | Protein-tyrosine-phosphatase, VANADATE ION | | Authors: | Shen, R.D, Hengge, A.C, Johnson, S.J. | | Deposit date: | 2020-12-11 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Single Residue on the WPD-Loop Affects the pH Dependency of Catalysis in Protein Tyrosine Phosphatases.
Jacs Au, 1, 2021
|
|
4UUN
 
 | |
7OOX
 
 | | Crystal structure of PIM1 in complex with ARC-3126 | | Descriptor: | 1,2-ETHANEDIOL, Inhibitor ARC-3126, PHOSPHATE ION, ... | | Authors: | Chaikuad, A, Dixon-Clarke, S.E, Nonga, O.E, Uri, A, Bullock, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-05-28 | | Release date: | 2021-08-04 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal Structure-Guided Design of Bisubstrate Inhibitors and Photoluminescent Probes for Protein Kinases of the PIM Family.
Molecules, 26, 2021
|
|
5LW9
 
 | | Crystal structure of human JARID1B in complex with S40563a | | Descriptor: | 1,2-ETHANEDIOL, 8-[4-[2-[4-[3,5-bis(chloranyl)phenyl]piperidin-1-yl]ethyl]pyrazol-1-yl]-3~{H}-pyrido[3,4-d]pyrimidin-4-one, DIMETHYL SULFOXIDE, ... | | Authors: | Srikannathasan, V, Le Bihan, Y.V, Szykowska, A, Johansson, C, Gileadi, C, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Oppermann, U, Huber, K. | | Deposit date: | 2016-09-15 | | Release date: | 2016-09-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of human JARID1B in complex with S40563a
to be published
|
|
