2OG1
 
 | | Crystal Structure of BphD, a C-C hydrolase from Burkholderia xenovorans LB400 | | Descriptor: | 2-hydroxy-6-oxo-6-phenylhexa-2,4-dienoate hydrolase, ETHANOL, GLYCEROL, ... | | Authors: | Dai, S, Ke, J, Bolin, J.T. | | Deposit date: | 2007-01-04 | | Release date: | 2007-01-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Kinetic and structural insight into the mechanism of BphD, a C-C bond hydrolase from the biphenyl degradation pathway
Biochemistry, 45, 2006
|
|
1OUZ
 
 | | Crystal structure of a mutant IHF (BetaE44A) complexed with a variant H' Site (T44A) | | Descriptor: | 5'-D(*GP*CP*TP*TP*AP*TP*CP*AP*AP*TP*TP*TP*GP*TP*AP*GP*CP*AP*CP*C)-3', 5'-D(*GP*GP*CP*CP*AP*AP*AP*AP*AP*AP*GP*CP*AP*TP*T)-3', Integration Host Factor Alpha-subunit, ... | | Authors: | Lynch, T.W, Read, E.K, Mattis, A.N, Gardner, J.F, Rice, P.A. | | Deposit date: | 2003-03-25 | | Release date: | 2003-07-15 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Integration Host Factor: putting a twist on protein-DNA recognition
J.Mol.Biol., 330, 2003
|
|
3D7K
 
 | | Crystal structure of benzaldehyde lyase in complex with the inhibitor MBP | | Descriptor: | 3-[(4-amino-2-methylpyrimidin-5-yl)methyl]-2-{(S)-hydroxy[(R)-hydroxy(methoxy)phosphoryl]phenylmethyl}-5-(2-{[(R)-hydroxy(phosphonooxy)phosphoryl]oxy}ethyl)-4-methyl-1,3-thiazol-3-ium, Benzaldehyde lyase, CALCIUM ION | | Authors: | Brandt, G.S. | | Deposit date: | 2008-05-21 | | Release date: | 2008-07-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Probing the active center of benzaldehyde lyase with substitutions and the pseudosubstrate analogue benzoylphosphonic acid methyl ester
Biochemistry, 47, 2008
|
|
5XKN
 
 | | Crystal structure of plant receptor ERL2 in complexe with EPFL4 | | Descriptor: | EPIDERMAL PATTERNING FACTOR-like protein 4, LRR receptor-like serine/threonine-protein kinase ERL2 | | Authors: | Chai, J, Lin, G. | | Deposit date: | 2017-05-08 | | Release date: | 2019-01-23 | | Method: | X-RAY DIFFRACTION (3.651 Å) | | Cite: | A receptor-like protein acts as a specificity switch for the regulation of stomatal development.
Genes Dev., 31, 2017
|
|
3CH3
 
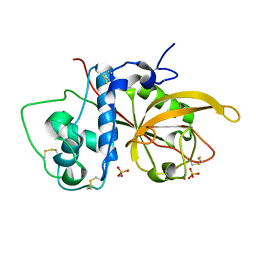 | | Crystal Structure Analysis of SERA5E from plasmodium falciparum | | Descriptor: | DIHYDROGENPHOSPHATE ION, POTASSIUM ION, Serine-repeat antigen protein | | Authors: | Smith, B.J, Malby, R.L, Colman, P.M, Clarke, O.B. | | Deposit date: | 2008-03-07 | | Release date: | 2008-03-25 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural Insights into the Protease-like Antigen Plasmodium falciparum SERA5 and Its Noncanonical Active-Site Serine
J.Mol.Biol., 392, 2009
|
|
4N64
 
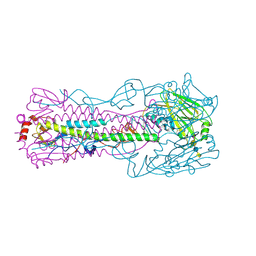 | |
3ANZ
 
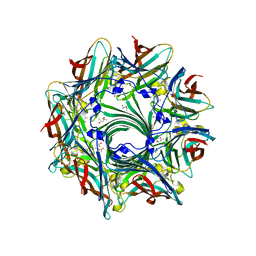 | | Crystal Structure of alpha-hemolysin | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETIC ACID, Alpha-hemolysin | | Authors: | Yamashita, K, Kawauchi, H, Tanaka, Y, Yao, M, Tanaka, I. | | Deposit date: | 2010-09-16 | | Release date: | 2011-06-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | 2-Methyl-2,4-pentanediol induces spontaneous assembly of staphylococcal alpha-hemolysin into heptameric pore structure
Protein Sci., 20, 2011
|
|
1RG8
 
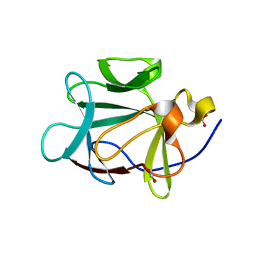 | |
4NBQ
 
 | | Structure of the polynucleotide phosphorylase (CBU_0852) from Coxiella burnetii | | Descriptor: | Polyribonucleotide nucleotidyltransferase, SULFATE ION | | Authors: | Rudolph, M.J, Cheung, J, Franklin, M.C, Cassidy, M, Gary, E, Burshteyn, F, Love, J. | | Deposit date: | 2013-10-23 | | Release date: | 2015-06-17 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.9138 Å) | | Cite: | Structural genomics for drug design against the pathogen Coxiella burnetii.
Proteins, 83, 2015
|
|
2PG7
 
 | | Crystal Structure of Human Microsomal P450 2A6 N297Q/I300V | | Descriptor: | Cytochrome P450 2A6, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sansen, S, Hsu, M.H, Stout, C.D, Johnson, E.F. | | Deposit date: | 2007-04-06 | | Release date: | 2007-07-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insight into the altered substrate specificity of human cytochrome P450 2A6 mutants.
Arch.Biochem.Biophys., 464, 2007
|
|
1R5Q
 
 | |
1R6Q
 
 | | ClpNS with fragments | | Descriptor: | ATP-dependent Clp protease ATP-binding subunit clpA, ATP-dependent Clp protease adaptor protein clpS, BIS-(2-HYDROXYETHYL)AMINO-TRIS(HYDROXYMETHYL)METHANE YTTRIUM, ... | | Authors: | Xia, D, Maurizi, M.R, Guo, F, Singh, S.K, Esser, L. | | Deposit date: | 2003-10-16 | | Release date: | 2005-02-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystallographic investigation of peptide binding sites in the N-domain of the ClpA chaperone.
J.Struct.Biol., 146, 2004
|
|
4HRT
 
 | | Scapharca tetrameric hemoglobin, unliganded | | Descriptor: | Globin-2 A chain, Hemoglobin B chain, PHOSPHATE ION, ... | | Authors: | Royer, W.E. | | Deposit date: | 2012-10-28 | | Release date: | 2012-12-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Tertiary and Quaternary Allostery in Tetrameric Hemoglobin from Scapharca inaequivalvis.
Biochemistry, 52, 2013
|
|
1RCQ
 
 | | The 1.45 A crystal structure of alanine racemase from a pathogenic bacterium, Pseudomonas aeruginosa, contains both internal and external aldimine forms | | Descriptor: | D-LYSINE, PYRIDOXAL-5'-PHOSPHATE, catabolic alanine racemase DadX | | Authors: | Le Magueres, P, Im, H, Dvorak, A, Strych, U, Benedik, M, Krause, K.L. | | Deposit date: | 2003-11-04 | | Release date: | 2004-06-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure at 1.45 A resolution of alanine racemase from a pathogenic bacterium, Pseudomonas aeruginosa, contains both internal and external aldimine forms.
Biochemistry, 42, 2003
|
|
4RKU
 
 | | Crystal structure of plant Photosystem I at 3 Angstrom resolution | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Mazor, Y, Borovikova, A, Greenberg, I, Nelson, N. | | Deposit date: | 2014-10-14 | | Release date: | 2015-02-04 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of plant Photosystem I at 3.1 Angstrom resolution
To be Published
|
|
4S21
 
 | | Crystal structure of the photosensory core module of bacteriophytochrome RPA3015 from R. palustris | | Descriptor: | BILIVERDINE IX ALPHA, Bacteriophytochrome (Light-regulated signal transduction histidine kinase), PhyB1 | | Authors: | Yang, X, Stojkovi, E.A, Ozarowski, W.B, Moffat, K. | | Deposit date: | 2015-01-17 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Light Signaling Mechanism of Two Tandem Bacteriophytochromes.
Structure, 23, 2015
|
|
5XA3
 
 | | Crystal Structure of P450BM3 with Benzyloxycarbonyl-L-prolyl-L-phenylalanine | | Descriptor: | Bifunctional cytochrome P450/NADPH-P450 reductase, DIMETHYL SULFOXIDE, PHENYLALANINE, ... | | Authors: | Shoji, O, Yanagisawa, S, Stanfield, J.K, Suzuki, K, Kasai, C, Cong, Z, Sugimoto, H, Shiro, Y, Watanabe, Y. | | Deposit date: | 2017-03-10 | | Release date: | 2018-02-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Direct Hydroxylation of Benzene to Phenol by Cytochrome P450BM3 Triggered by Amino Acid Derivatives.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
3AOH
 
 | | RNA polymerase-Gfh1 complex (Crystal type 1) | | Descriptor: | Anti-cleavage anti-GreA transcription factor Gfh1, DNA (5'-D(*GP*GP*TP*CP*TP*GP*TP*AP*TP*CP*AP*CP*GP*AP*GP*CP*CP*AP*CP*CP*GP*CP*CP*GP*CP*AP*T)-3'), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Tagami, S, Sekine, S, Kumarevel, T, Yamamoto, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2010-09-28 | | Release date: | 2010-12-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Crystal structure of bacterial RNA polymerase bound with a transcription inhibitor protein
Nature, 468, 2010
|
|
2PR6
 
 | |
2PRT
 
 | | Structure of the Wilms Tumor Suppressor Protein Zinc Finger Domain Bound to DNA | | Descriptor: | DNA (5'-D(*CP*AP*GP*AP*CP*GP*CP*CP*CP*CP*CP*GP*CP*G)-3'), DNA (5'-D(*CP*GP*CP*GP*GP*GP*GP*GP*CP*GP*TP*CP*TP*G)-3'), Wilms tumor 1, ... | | Authors: | Stoll, R, Lee, B.M, Debler, E.W, Laity, J.H, Wilson, I.A, Dyson, H.J, Wright, P.E. | | Deposit date: | 2007-05-04 | | Release date: | 2008-03-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structure of the Wilms tumor suppressor protein zinc finger domain bound to DNA
J.Mol.Biol., 372, 2007
|
|
4QWJ
 
 | | yCP beta5-A49T-mutant in complex with carfilzomib | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Huber, E.M, Heinemeyer, W, Groll, M. | | Deposit date: | 2014-07-16 | | Release date: | 2015-02-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Bortezomib-Resistant Mutant Proteasomes: Structural and Biochemical Evaluation with Carfilzomib and ONX 0914.
Structure, 23, 2015
|
|
1R1T
 
 | | Crystal structure of the cyanobacterial metallothionein repressor SmtB in the apo-form | | Descriptor: | Transcriptional repressor smtB | | Authors: | Eicken, C, Pennella, M.A, Chen, X, Koshlap, K.M, VanZile, M.L, Sacchettini, J.C, Giedroc, D.P. | | Deposit date: | 2003-09-25 | | Release date: | 2004-05-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A metal-ligand-mediated intersubunit allosteric switch in related SmtB/ArsR zinc sensor proteins.
J.Mol.Biol., 333, 2003
|
|
4QZ2
 
 | | yCP beta5-M45I mutant in complex with the epoxyketone inhibitor ONX 0914 | | Descriptor: | 1,2,4-trideoxy-4-methyl-2-{[N-(morpholin-4-ylacetyl)-L-alanyl-O-methyl-L-tyrosyl]amino}-1-phenyl-D-xylitol, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Huber, E.M, Heinemeyer, W, Groll, M. | | Deposit date: | 2014-07-27 | | Release date: | 2015-02-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Bortezomib-Resistant Mutant Proteasomes: Structural and Biochemical Evaluation with Carfilzomib and ONX 0914.
Structure, 23, 2015
|
|
5YXD
 
 | | A ligand F binding to FXR | | Descriptor: | Bile acid receptor, Peptide from Nuclear receptor coactivator, ethyl methyl 4-(2,3-dichlorophenyl)-2,6-dimethylpyridine-3,5-dicarboxylate | | Authors: | Yi, L, Yong, L. | | Deposit date: | 2017-12-05 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | A ligand F binding to FXR
To Be Published
|
|
2PZ5
 
 | |
