9R2J
 
 | | Crystal structure of human MAO B in complex with (E)-3-(4-nitrophenyl)-1-(3-(trifluoromethyl)phenyl)prop-2-en-1-one (chalcone inhibitor, 4a) | | Descriptor: | (~{E})-3-(4-nitrophenyl)-1-[3-(trifluoromethyl)phenyl]prop-2-en-1-one, Amine oxidase [flavin-containing] B, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Marchese, S, Binda, C. | | Deposit date: | 2025-04-30 | | Release date: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design, synthesis, and biological evaluation of chalcone derivatives as selective Monoamine Oxidase-B inhibitors with potential neuroprotective effects.
Eur.J.Med.Chem., 298, 2025
|
|
8CVF
 
 | | Structure of L289F Hyoscyamine 6-beta Hydroxylase in complex with vanadyl, succinate, and hyoscyamine | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, Hyoscyamine 6-beta-hydroxylase, ... | | Authors: | Wenger, E.S, Boal, A.K, Bollinger, J.M, Krebs, C. | | Deposit date: | 2022-05-18 | | Release date: | 2023-11-22 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (1.532 Å) | | Cite: | Optimized Substrate Positioning Enables Switches in the C-H Cleavage Site and Reaction Outcome in the Hydroxylation-Epoxidation Sequence Catalyzed by Hyoscyamine 6 beta-Hydroxylase.
J.Am.Chem.Soc., 146, 2024
|
|
8CVE
 
 | | Structure of L289F Hyoscyamine 6-beta Hydroxylase in complex with iron, 2-oxoglutarate, and hyoscyamine | | Descriptor: | 1,2-ETHANEDIOL, 2-OXOGLUTARIC ACID, FE (II) ION, ... | | Authors: | Wenger, E.S, Boal, A.K, Bollinger, J.M, Krebs, C. | | Deposit date: | 2022-05-18 | | Release date: | 2023-11-22 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Optimized Substrate Positioning Enables Switches in the C-H Cleavage Site and Reaction Outcome in the Hydroxylation-Epoxidation Sequence Catalyzed by Hyoscyamine 6 beta-Hydroxylase.
J.Am.Chem.Soc., 146, 2024
|
|
5LI6
 
 | | Crystal structure of Mycobacterium tuberculosis CYP126A1 in complex with N-isopropyl-N-((3-(4-methoxyphenyl)-1,2,4-oxadiazol-5-yl)methyl)-2-(4-nitrophenyl)acetamide | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Putative cytochrome P450 126, ~{N}-[[3-(4-methoxyphenyl)-1,2,4-oxadiazol-5-yl]methyl]-2-(4-nitrophenyl)-~{N}-propan-2-yl-ethanamide | | Authors: | Levy, C, Munro, A.W, Leys, D. | | Deposit date: | 2016-07-14 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Characterization and Ligand/Inhibitor Identification Provide Functional Insights into the Mycobacterium tuberculosis Cytochrome P450 CYP126A1.
J. Biol. Chem., 292, 2017
|
|
5L3E
 
 | | LSD1-CoREST1 in complex with quinazoline-derivative reversible inhibitor | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Lysine-specific histone demethylase 1A, N~4~-(1-benzylpiperidin-4-yl)-N~2~-[3-(dimethylamino)propyl]-6,7-dimethoxyquinazoline-2,4-diamine, ... | | Authors: | Speranzini, V, Rotili, D, Ciossani, G, Pilotto, S, Forgione, M, Lucidi, A, Forneris, F, Velankar, S, Mai, A, Mattevi, A. | | Deposit date: | 2016-04-10 | | Release date: | 2016-09-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Polymyxins and quinazolines are LSD1/KDM1A inhibitors with unusual structural features.
Sci Adv, 2, 2016
|
|
9H44
 
 | | Jumonji domain-containing protein 2B with crown ether and crystallization epitope mutations L916G:R917A:A918D | | Descriptor: | 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, CHLORIDE ION, Lysine-specific demethylase 4B | | Authors: | Fairhead, M, Strain-Damerell, C, Ye, M, Mackinnon, S.R, Pinkas, D, MacLean, E.M, Koekemoer, L, Damerell, D, Krojer, T, Yue, W, Burgess-Brown, N, Marsden, B, von Delft, F, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-10-17 | | Release date: | 2024-12-04 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | A fast, parallel method for efficiently exploring crystallization behaviour of large numbers of protein variants
To Be Published
|
|
4U83
 
 | |
8OH8
 
 | | Crystal structure of the cysteine-rich Gallus gallus urate oxidase in complex with the 8-azaxanthine inhibitor under reducing conditions (space group P 21 21 21) | | Descriptor: | 1,2-ETHANEDIOL, 8-AZAXANTHINE, CHLORIDE ION, ... | | Authors: | Di Palma, M, Chegkazi, M, Bui, S, Mori, G, Percudani, R, Steiner, R.A. | | Deposit date: | 2023-03-20 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Cysteine Enrichment Mediates Co-Option of Uricase in Reptilian Skin and Transition to Uricotelism.
Mol.Biol.Evol., 40, 2023
|
|
8OIH
 
 | | Crystal structure of the cysteine-rich Gallus gallus urate oxidase in complex with the 8-azaxanthine inhibitor under oxidising conditions (space group C 2 2 21) | | Descriptor: | 1,2-ETHANEDIOL, 8-AZAXANTHINE, BROMIDE ION, ... | | Authors: | Di Palma, M, Chegkazi, M, Bui, S, Mori, G, Percudani, R, Steiner, R.A. | | Deposit date: | 2023-03-22 | | Release date: | 2024-01-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Cysteine Enrichment Mediates Co-Option of Uricase in Reptilian Skin and Transition to Uricotelism.
Mol.Biol.Evol., 40, 2023
|
|
6VBN
 
 | | Crystal Structure of hTDO2 bound to inhibitor GNE1 | | Descriptor: | 1,5-anhydro-2,3-dideoxy-3-[(5S)-5H-imidazo[5,1-a]isoindol-5-yl]-D-threo-pentitol, PROTOPORPHYRIN IX CONTAINING FE, Tryptophan 2,3-dioxygenase | | Authors: | Harris, S.F, Oh, A. | | Deposit date: | 2019-12-19 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | Implementation of the CYP Index for the Design of Selective Tryptophan-2,3-dioxygenase Inhibitors.
Acs Med.Chem.Lett., 11, 2020
|
|
6G5X
 
 | | Crystal Structure of KDM4A with compound YP-02-145 | | Descriptor: | 1,2-ETHANEDIOL, 2-(3-methyl-5-oxidanylidene-4-phenyl-4~{H}-pyrazol-1-yl)-3~{H}-benzimidazole-5-carboxylic acid, CITRIC ACID, ... | | Authors: | Malecki, P.H, Carter, D.M, Gohlke, U, Specker, E, Nazare, M, Weiss, M.S, Heinemann, U. | | Deposit date: | 2018-03-30 | | Release date: | 2019-04-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Enhanced Properties of a Benzimidazole Benzylpyrazole Lysine Demethylase Inhibitor: Mechanism-of-Action, Binding Site Analysis, and Activity in Cellular Models of Prostate Cancer.
J.Med.Chem., 64, 2021
|
|
5LI8
 
 | | Crystal structure of Mycobacterium tuberculosis CYP126A1 in complex with ketoconazole | | Descriptor: | 1-acetyl-4-(4-{[(2R,4S)-2-(2,4-dichlorophenyl)-2-(1H-imidazol-1-ylmethyl)-1,3-dioxolan-4-yl]methoxy}phenyl)piperazine, PROTOPORPHYRIN IX CONTAINING FE, Putative cytochrome P450 126 | | Authors: | Levy, C, Munro, A.W, Leys, D. | | Deposit date: | 2016-07-14 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural Characterization and Ligand/Inhibitor Identification Provide Functional Insights into the Mycobacterium tuberculosis Cytochrome P450 CYP126A1.
J. Biol. Chem., 292, 2017
|
|
4QOJ
 
 | | CRYSTAL STRUCTURE OF FMN QUINONE REDUCTASE 2 IN COMPLEX WITH RESVERATROL AT 1.85A | | Descriptor: | FLAVIN MONONUCLEOTIDE, RESVERATROL, Ribosyldihydronicotinamide dehydrogenase [quinone], ... | | Authors: | Serriere, J, Boutin, J.A, Isabet, T, Antoine, M, Ferry, G. | | Deposit date: | 2014-06-20 | | Release date: | 2015-08-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | CRYSTAL STRUCTURE OF FMN QUINONE REDUCTASE 2 IN COMPLEX WITH RESVERATROL AT 1.85A
To be Published
|
|
4QOI
 
 | | Crystal structure of FMN quinone reductase 2 in complex with melatonin at 1.55A | | Descriptor: | FLAVIN MONONUCLEOTIDE, N-[2-(5-methoxy-1H-indol-3-yl)ethyl]acetamide, Ribosyldihydronicotinamide dehydrogenase [quinone], ... | | Authors: | Serriere, J, Boutin, J.A, Isabet, T, Antoine, M, Ferry, G. | | Deposit date: | 2014-06-20 | | Release date: | 2015-07-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of FMN quinone reductase 2 in complex with melatonin at 1.55A
To be Published
|
|
4QOG
 
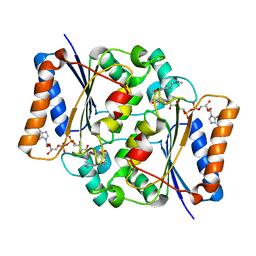 | | Crystal structure of fad quinone reductase 2 in complex with melatonin at 1.4A | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, N-[2-(5-methoxy-1H-indol-3-yl)ethyl]acetamide, Ribosyldihydronicotinamide dehydrogenase [quinone], ... | | Authors: | Serriere, J, Boutin, J.A, Isabet, T, Antoine, M, Ferry, G. | | Deposit date: | 2014-06-20 | | Release date: | 2015-07-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of fad quinone reductase 2 in complex with melatonin at 1.4A
To be Published
|
|
8AE8
 
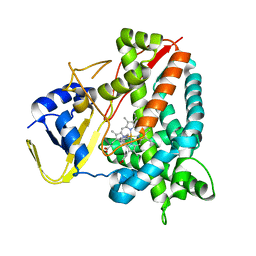 | |
6WXZ
 
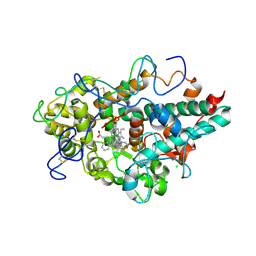 | | CRYSTAL STRUCTURE OF MYELOPEROXIDASE SUBFORM C (MPO) COMPLEX WITH Compound-29 A.K.A 7-(1,2-DIPHENYLETHYL)-1H-[1,2,3]TRIAZOLO[4,5-B]PYRIDIN-5-AMINE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 7-[(1R)-1,2-diphenylethyl]-3H-[1,2,3]triazolo[4,5-b]pyridin-5-amine, CALCIUM ION, ... | | Authors: | Khan, J.A. | | Deposit date: | 2020-05-12 | | Release date: | 2020-10-14 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.226 Å) | | Cite: | Discovery and structure activity relationships of 7-benzyl triazolopyridines as stable, selective, and reversible inhibitors of myeloperoxidase.
Bioorg.Med.Chem., 28, 2020
|
|
8RQZ
 
 | | Crystal structure of Molybdenum bispyranopterin guanine dinucleotide formate dehydrogenases ForCE1 from Bacillus subtilis | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Cherrier, M.V, Arnoux, P, Martin, L, Nicolet, Y, Schoehn, G, Legrand, P, Broc, M, Seduk, F, Brasseur, G, Arias-Cartin, R, Magalon, A, Walburger, A, Uzel, A, Guigliarelli, B, Grimaldi, S, Pierrel, F, Mate, M. | | Deposit date: | 2024-01-22 | | Release date: | 2025-08-06 | | Last modified: | 2025-09-03 | | Method: | X-RAY DIFFRACTION (2.694 Å) | | Cite: | A scaffold for quinone channeling between membrane and soluble bacterial oxidoreductases.
Nat.Struct.Mol.Biol., 2025
|
|
6QYH
 
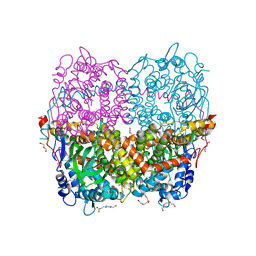 | | Structure of Apo HPAB from E.coli | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 4-hydroxyphenylacetate 3-monooxygenase oxygenase component, CALCIUM ION, ... | | Authors: | Pecqueur, L, Lombard, M, Deng, Y, Fontecave, M. | | Deposit date: | 2019-03-09 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural and Functional Characterization of 4-Hydroxyphenylacetate 3-Hydroxylase from Escherichia coli.
Chembiochem, 21, 2020
|
|
7R1I
 
 | | Crystal structure of CYP125 from Mycobacterium tuberculosis in complex with an inhibitor | | Descriptor: | CHLORIDE ION, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION, ... | | Authors: | Snee, M, Katariya, M, Levy, C, Leys, D. | | Deposit date: | 2022-02-03 | | Release date: | 2023-02-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structure Based Discovery of Inhibitors of CYP125 and CYP142 from Mycobacterium tuberculosis.
Chemistry, 29, 2023
|
|
8AVH
 
 | | Crystal structure of IsdG from Bacillus cereus | | Descriptor: | 1,3-PROPANDIOL, Heme-degrading monooxygenase | | Authors: | Andersen, H.K, Hammerstad, M, Hersleth, H.-P. | | Deposit date: | 2022-08-26 | | Release date: | 2023-07-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Functional Diversity of Homologous Oxidoreductases-Tuning of Substrate Specificity by a FAD-Stacking Residue for Iron Acquisition and Flavodoxin Reduction.
Antioxidants, 12, 2023
|
|
6H0X
 
 | | Crystal Structure of KDM4D with tetrazolylhydrazide ligand AA040 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysine-specific demethylase 4D, ... | | Authors: | Malecki, P.H, Weiss, M.S, Heinemann, U, Link, A. | | Deposit date: | 2018-07-10 | | Release date: | 2020-01-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal Structure of KDM4D with tetrazolylhydrazide ligand AA040
To be published
|
|
6H11
 
 | | Crystal Structure of KDM4D with tetrazolylhydrazide ligand AA028 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysine-specific demethylase 4D, ... | | Authors: | Malecki, P.H, Weiss, M.S, Heinemann, U, Link, A. | | Deposit date: | 2018-07-10 | | Release date: | 2020-01-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.516 Å) | | Cite: | Crystal Structure of KDM4D with tetrazolylhydrazide ligand AA028
To be published
|
|
5NAE
 
 | |
6I34
 
 | | Crystal structure of Neanderthal glycine decarboxylase (P-protein) | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Van Laer, B, Kapp, U, Leonard, G, Mueller-Dieckmann, C. | | Deposit date: | 2018-11-05 | | Release date: | 2019-11-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insights in human glycine decarboxylase and comparison with the Neanderthal variant
To Be Published
|
|
