7L37
 
 | | T4 Lysozyme L99A - Apo - RT | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Fischer, M, Bradford, S.Y.C. | | Deposit date: | 2020-12-17 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.439 Å) | | Cite: | Temperature artifacts in protein structures bias ligand-binding predictions.
Chem Sci, 12, 2021
|
|
7L3B
 
 | | T4 Lysozyme L99A - iodobenzene - RT | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Fischer, M, Bradford, S.Y.C. | | Deposit date: | 2020-12-17 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Temperature artifacts in protein structures bias ligand-binding predictions.
Chem Sci, 12, 2021
|
|
7L3G
 
 | | T4 Lysozyme L99A - 4-iodotoluene - cryo | | Descriptor: | 1-iodo-4-methylbenzene, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, ... | | Authors: | Fischer, M, Bradford, S.Y.C. | | Deposit date: | 2020-12-17 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Temperature artifacts in protein structures bias ligand-binding predictions.
Chem Sci, 12, 2021
|
|
7L3H
 
 | | T4 Lysozyme L99A - ethylbenzene - RT | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Fischer, M, Bradford, S.Y.C. | | Deposit date: | 2020-12-17 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Temperature artifacts in protein structures bias ligand-binding predictions.
Chem Sci, 12, 2021
|
|
7L3F
 
 | | T4 Lysozyme L99A - 4-iodotoluene - RT | | Descriptor: | 1-iodo-4-methylbenzene, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Fischer, M, Bradford, S.Y.C. | | Deposit date: | 2020-12-17 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Temperature artifacts in protein structures bias ligand-binding predictions.
Chem Sci, 12, 2021
|
|
7L3K
 
 | | T4 Lysozyme L99A - benzylacetate - cryo | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Fischer, M, Bradford, S.Y.C. | | Deposit date: | 2020-12-17 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | Temperature artifacts in protein structures bias ligand-binding predictions.
Chem Sci, 12, 2021
|
|
7L3C
 
 | | T4 Lysozyme L99A - o-xylene - RT | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Fischer, M, Bradford, S.Y.C. | | Deposit date: | 2020-12-17 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Temperature artifacts in protein structures bias ligand-binding predictions.
Chem Sci, 12, 2021
|
|
7L3E
 
 | | T4 Lysozyme L99A - 3-iodotoluene - cryo | | Descriptor: | 1-iodo-3-methylbenzene, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, ... | | Authors: | Fischer, M, Bradford, S.Y.C. | | Deposit date: | 2020-12-17 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Temperature artifacts in protein structures bias ligand-binding predictions.
Chem Sci, 12, 2021
|
|
7L3D
 
 | | T4 Lysozyme L99A - 3-iodotoluene - RT | | Descriptor: | 1-iodo-3-methylbenzene, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, ... | | Authors: | Fischer, M, Bradford, S.Y.C. | | Deposit date: | 2020-12-17 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Temperature artifacts in protein structures bias ligand-binding predictions.
Chem Sci, 12, 2021
|
|
8IUS
 
 | |
7L3I
 
 | | T4 Lysozyme L99A - propylbenzene - RT | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, Endolysin, ... | | Authors: | Fischer, M, Bradford, S.Y.C. | | Deposit date: | 2020-12-17 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Temperature artifacts in protein structures bias ligand-binding predictions.
Chem Sci, 12, 2021
|
|
8IUT
 
 | |
8IUU
 
 | |
8IJG
 
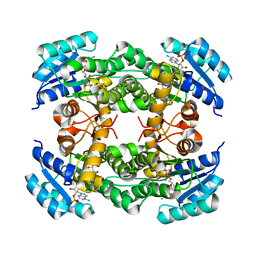 | | Crystal structure of alcohol dehydrogenase M5 from Burkholderia gladioli with NADP | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Putative short-chain dehydrogenases/reductase family protein | | Authors: | Han, X, Mei, Z.L, Liu, W.D, Sun, Z.T, Ma, J.A. | | Deposit date: | 2023-02-27 | | Release date: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystal structure of alcohol dehydrogenase from Burkholderia gladioli with NADP
To Be Published
|
|
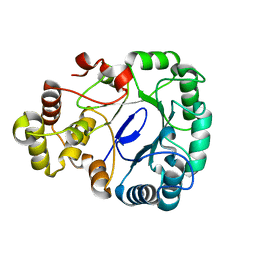
8IKU
 
 | |
7L4X
 
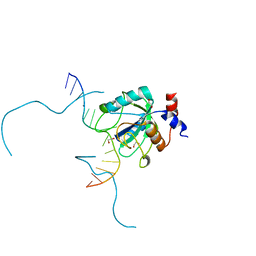 | |
7L4Y
 
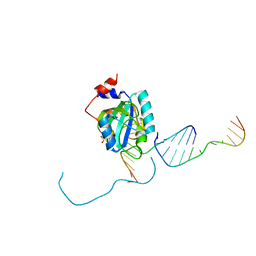 | |
7JXV
 
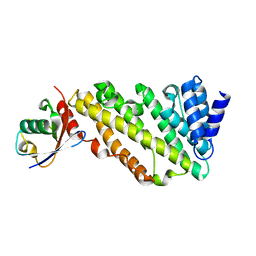 | | ANTH domain of CALM (clathrin-assembly lymphoid myeloid leukemia protein) bound to ubiquitin | | Descriptor: | Phosphatidylinositol-binding clathrin assembly protein, Ubiquitin | | Authors: | Pashkova, N, Gakhar, L, Schnicker, N.J, Piper, R.C. | | Deposit date: | 2020-08-28 | | Release date: | 2021-08-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | ANTH domains within CALM, HIP1R, and Sla2 recognize ubiquitin internalization signals.
Elife, 10, 2021
|
|
3BDZ
 
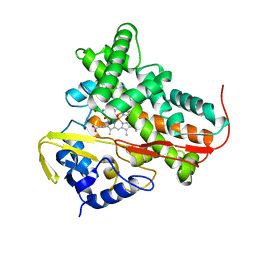 | | The Role of Asn 242 in P450cin | | Descriptor: | MALONATE ION, P450cin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Meharenna, Y.T, Poulos, T.L. | | Deposit date: | 2007-11-15 | | Release date: | 2008-02-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The critical role of substrate-protein hydrogen bonding in the control of regioselective hydroxylation in p450cin
J.Biol.Chem., 283, 2008
|
|
3BHI
 
 | |
8IVY
 
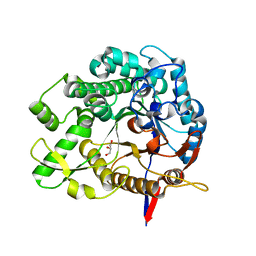 | | Beta-Glucosidase BglA mutant E166Q in complex with glucose | | Descriptor: | Beta-glucosidase, GLYCEROL, beta-D-glucopyranose | | Authors: | Dong, S, Xiao, Y, Feng, Y. | | Deposit date: | 2023-03-29 | | Release date: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Key roles of beta-glucosidase BglA for the catabolism of both laminaribiose and cellobiose in the lignocellulolytic bacterium Clostridium thermocellum.
Int.J.Biol.Macromol., 250, 2023
|
|
8J8Q
 
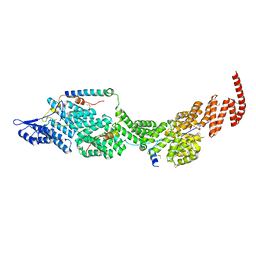 | | Structure of the four-component Paf1 complex from Saccharomyces eubayanus | | Descriptor: | CDC73-like protein, CTR9-like protein, PAF1-like protein, ... | | Authors: | Wang, Z, Qin, Y, Zhou, Y, Cao, Y. | | Deposit date: | 2023-05-02 | | Release date: | 2023-05-24 | | Last modified: | 2023-06-14 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Structural Basis of the Transcriptional Elongation Factor Paf1 Core Complex from Saccharomyces eubayanus .
Int J Mol Sci, 24, 2023
|
|
8JBZ
 
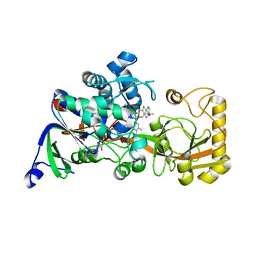 | | Crystal structure of 3-ketosteroid delta1-dehydrogenase from Rhodococcus erythropolis SQ1 in complex with 4-androstadiene-3,17- dione | | Descriptor: | 3-ketosteroid dehydrogenase, 4-ANDROSTENE-3-17-DIONE, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Hu, Y.L, Li, X, Cheng, X.Y, Song, S.K, Su, Z.D. | | Deposit date: | 2023-05-10 | | Release date: | 2023-05-31 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.079 Å) | | Cite: | Crystal structure of 3-ketosteroid delta1-dehydrogenase from Rhodococcus erythropolis SQ1 in complex with 4-androstadiene-3,17- dione
To Be Published
|
|
8J8P
 
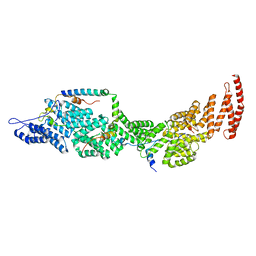 | | Structure of the four-component Paf1 complex from Saccharomyces eubayanus | | Descriptor: | CDC73-like protein, CTR9-like protein, PAF1-like protein, ... | | Authors: | Wang, Z, Qin, Y, Zhou, Y, Cao, Y. | | Deposit date: | 2023-05-02 | | Release date: | 2023-05-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis of the Transcriptional Elongation Factor Paf1 Core Complex from Saccharomyces eubayanus .
Int J Mol Sci, 24, 2023
|
|
8IVJ
 
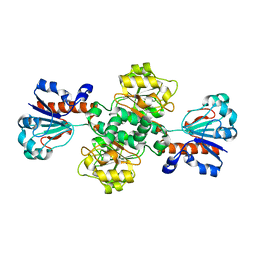 | |
