3H5R
 
 | | Crystal structure of E. coli MccB + Succinimide | | Descriptor: | MccB protein, Microcin C7 analog, SULFATE ION, ... | | Authors: | Regni, C.A, Roush, R.F, Miller, D, Nourse, A, Walsh, C.T, Schulman, B.A. | | Deposit date: | 2009-04-22 | | Release date: | 2009-06-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | How the MccB bacterial ancestor of ubiquitin E1 initiates biosynthesis of the microcin C7 antibiotic.
Embo J., 28, 2009
|
|
5FWL
 
 | | Atomic cryoEM structure of Hsp90-Cdc37-Cdk4 complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CYCLIN-DEPENDENT KINASE 4, HEAT SHOCK PROTEIN HSP 90 BETA, ... | | Authors: | Verba, K.A, Wang, R.Y.R, Arakawa, A, Liu, Y, Yokoyama, S, Agard, D.A. | | Deposit date: | 2016-02-18 | | Release date: | 2016-07-06 | | Last modified: | 2019-10-23 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Atomic Structure of Hsp90-Cdc37-Cdk4 Reveals that Hsp90 Traps and Stabilizes an Unfolded Kinase.
Science, 352, 2016
|
|
5FWM
 
 | | Atomic cryoEM structure of Hsp90-Cdc37-Cdk4 complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CYCLIN-DEPENDENT KINASE 4, HEAT SHOCK PROTEIN HSP 90 BETA, ... | | Authors: | Verba, K.A, Wang, R.Y.R, Arakawa, A, Liu, Y, Yokoyama, S, Agard, D.A. | | Deposit date: | 2016-02-18 | | Release date: | 2016-07-06 | | Last modified: | 2019-10-23 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Atomic Structure of Hsp90-Cdc37-Cdk4 Reveals that Hsp90 Traps and Stabilizes an Unfolded Kinase.
Science, 352, 2016
|
|
5FWP
 
 | | Atomic cryoEM structure of Hsp90-Cdc37-Cdk4 complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CYCLIN-DEPENDENT KINASE 4, HEAT SHOCK PROTEIN HSP 90 BETA, ... | | Authors: | Verba, K.A, Wang, R.Y.R, Arakawa, A, Liu, Y, Yokoyama, S, Agard, D.A. | | Deposit date: | 2016-02-18 | | Release date: | 2016-10-26 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (7.2 Å) | | Cite: | Atomic Structure of Hsp90:Cdc37:Cdk4 Reveals that Hsp90 Traps and Stabilizes an Unfolded Kinase.
Science, 352, 2016
|
|
3VH8
 
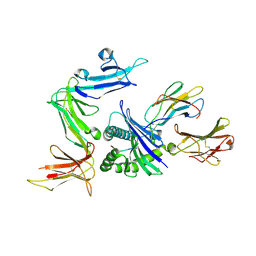 | | KIR3DL1 in complex with HLA-B*5701 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Vivian, J.P, Rossjohn, J. | | Deposit date: | 2011-08-24 | | Release date: | 2011-10-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Killer cell immunoglobulin-like receptor 3DL1-mediated recognition of human leukocyte antigen B
Nature, 479, 2011
|
|
5FWK
 
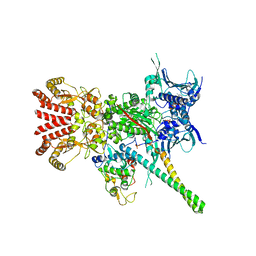 | | Atomic cryoEM structure of Hsp90-Cdc37-Cdk4 complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CYCLIN-DEPENDENT KINASE 4, HEAT SHOCK PROTEIN HSP 90 BETA, ... | | Authors: | Verba, K.A, Wang, R.Y.R, Arakawa, A, Liu, Y, Yokoyama, S, Agard, D.A. | | Deposit date: | 2016-02-17 | | Release date: | 2016-07-06 | | Last modified: | 2019-08-21 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Atomic Structure of Hsp90-Cdc37-Cdk4 Reveals that Hsp90 Traps and Stabilizes an Unfolded Kinase.
Science, 352, 2016
|
|
3H9G
 
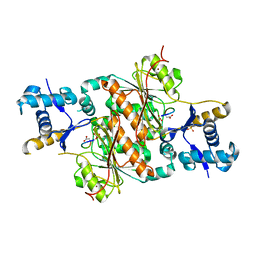 | | Crystal structure of E. coli MccB + MccA-N7isoASN | | Descriptor: | MccB protein, Microcin C7 analog, SULFATE ION, ... | | Authors: | Regni, C.A, Roush, R.F, Miller, D, Nourse, A, Walsh, C.T, Schulman, B.A. | | Deposit date: | 2009-04-30 | | Release date: | 2009-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | How the MccB bacterial ancestor of ubiquitin E1 initiates biosynthesis of the microcin C7 antibiotic.
Embo J., 28, 2009
|
|
3H5A
 
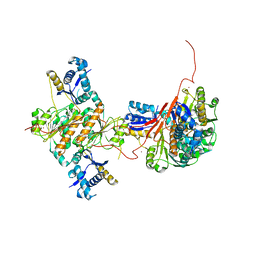 | | Crystal structure of E. coli MccB | | Descriptor: | MccB protein, ZINC ION | | Authors: | Regni, C.A, Roush, R.F, Miller, D, Nourse, A, Walsh, C.T, Schulman, B.A. | | Deposit date: | 2009-04-21 | | Release date: | 2009-06-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | How the MccB bacterial ancestor of ubiquitin E1 initiates biosynthesis of the microcin C7 antibiotic.
Embo J., 28, 2009
|
|
3H9Q
 
 | | Crystal structure of E. coli MccB + SeMet MccA | | Descriptor: | MccB protein, Microcin C7 ANALOG, SULFATE ION, ... | | Authors: | Regni, C.A, Roush, R.F, Miller, D, Nourse, A, Walsh, C.T, Schulman, B.A. | | Deposit date: | 2009-04-30 | | Release date: | 2009-06-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | How the MccB bacterial ancestor of ubiquitin E1 initiates biosynthesis of the microcin C7 antibiotic.
Embo J., 28, 2009
|
|
3H9J
 
 | | Crystal structure of E. coli MccB + AMPCPP + SeMeT MccA | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, MccB protein, Microcin C7 ANALOG, ... | | Authors: | Regni, C.A, Roush, R.F, Miller, D, Nourse, A, Walsh, C.T, Schulman, B.A. | | Deposit date: | 2009-04-30 | | Release date: | 2009-06-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | How the MccB bacterial ancestor of ubiquitin E1 initiates biosynthesis of the microcin C7 antibiotic.
Embo J., 28, 2009
|
|
3H5N
 
 | | Crystal structure of E. coli MccB + ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, MccB protein, ... | | Authors: | Regni, C.A, Roush, R.F, Miller, D, Nourse, A, Walsh, C.T, Schulman, B.A. | | Deposit date: | 2009-04-22 | | Release date: | 2009-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | How the MccB bacterial ancestor of ubiquitin E1 initiates biosynthesis of the microcin C7 antibiotic.
Embo J., 28, 2009
|
|
3V1Y
 
 | | Crystal structures of glyceraldehyde-3-phosphate dehydrogenase complexes with NAD | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase, cytosolic, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Tien, Y.C, Chuankhayan, P, Lin, Y.H, Chang, S.L, Chen, C.J. | | Deposit date: | 2011-12-10 | | Release date: | 2012-11-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal structures of rice (Oryza sativa) glyceraldehyde-3-phosphate dehydrogenase complexes with NAD and sulfate suggest involvement of Phe37 in NAD binding for catalysis
Plant Mol.Biol., 80, 2012
|
|
3WMG
 
 | | Crystal structure of an inward-facing eukaryotic ABC multidrug transporter G277V/A278V/A279V mutant in complex with an cyclic peptide inhibitor, aCAP | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ATP-binding cassette, sub-family B, ... | | Authors: | Kodan, A, Yamaguchi, T, Nakatsu, T, Sakiyama, K, Hipolito, C.J, Fujioka, A, Hirokane, R, Ikeguchi, K, Watanabe, B, Hirtake, J, Kimura, Y, Suga, H, Ueda, K, Kato, H. | | Deposit date: | 2013-11-18 | | Release date: | 2014-04-30 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for gating mechanisms of a eukaryotic P-glycoprotein homolog.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3WME
 
 | | Crystal structure of an inward-facing eukaryotic ABC multidrug transporter | | Descriptor: | ATP-binding cassette, sub-family B, member 1, ... | | Authors: | Kodan, A, Yamaguchi, T, Nakatsu, T, Kato, H. | | Deposit date: | 2013-11-18 | | Release date: | 2014-03-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.751 Å) | | Cite: | Structural basis for gating mechanisms of a eukaryotic P-glycoprotein homolog.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3WMF
 
 | | Crystal structure of an inward-facing eukaryotic ABC multitrug transporter G277V/A278V/A279V mutant | | Descriptor: | ATP-binding cassette, sub-family B, member 1, ... | | Authors: | Kodan, A, Yamaguchi, T, Nakatsu, T, Kato, H. | | Deposit date: | 2013-11-18 | | Release date: | 2014-03-19 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for gating mechanisms of a eukaryotic P-glycoprotein homolog.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3OCX
 
 | | Structure of Recombinant Haemophilus influenzae e(P4) Acid Phosphatase mutant D66N complexed with 2'-AMP | | Descriptor: | ADENOSINE-2'-MONOPHOSPHATE, Lipoprotein E, MAGNESIUM ION | | Authors: | Singh, H, Schuermann, J, Reilly, T, Calcutt, M, Tanner, J. | | Deposit date: | 2010-08-10 | | Release date: | 2010-10-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Recognition of nucleoside monophosphate substrates by Haemophilus influenzae class C acid phosphatase.
J.Mol.Biol., 404, 2010
|
|
3OCW
 
 | | Structure of Recombinant Haemophilus influenzae e(P4) Acid Phosphatase mutant D66N complexed with 3'-AMP | | Descriptor: | Lipoprotein E, MAGNESIUM ION, [(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-4-hydroxy-2-(hydroxymethyl)oxolan-3-yl] dihydrogen phosphate | | Authors: | Singh, H, Schuermann, J, Reilly, T, Calcutt, M, Tanner, J. | | Deposit date: | 2010-08-10 | | Release date: | 2010-10-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Recognition of nucleoside monophosphate substrates by Haemophilus influenzae class C acid phosphatase.
J.Mol.Biol., 404, 2010
|
|
4NM7
 
 | | Crystal structure of GSK-3/Axin complex bound to phosphorylated Wnt receptor LRP6 e-motif | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, ADENOSINE-5'-DIPHOSPHATE, Axin-1, ... | | Authors: | Stamos, J.L, Chu, M.L.-H, Enos, M.D, Shah, N, Weis, W.I. | | Deposit date: | 2013-11-14 | | Release date: | 2014-03-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of GSK-3 inhibition by N-terminal phosphorylation and by the Wnt receptor LRP6.
Elife, 3, 2014
|
|
4NU1
 
 | | Crystal structure of a transition state mimic of the GSK-3/Axin complex bound to phosphorylated N-terminal auto-inhibitory pS9 peptide | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, Axin-1, ... | | Authors: | Chu, M.L.-H, Stamos, J.L, Enos, M.D, Shah, N, Weis, W.I. | | Deposit date: | 2013-12-03 | | Release date: | 2014-03-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of GSK-3 inhibition by N-terminal phosphorylation and by the Wnt receptor LRP6.
Elife, 3, 2014
|
|
4NM5
 
 | | Crystal structure of GSK-3/Axin complex bound to phosphorylated Wnt receptor LRP6 c-motif | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Axin-1, CHLORIDE ION, ... | | Authors: | Stamos, J.L, Chu, M.L.-H, Enos, M.D, Shah, N, Weis, W.I. | | Deposit date: | 2013-11-14 | | Release date: | 2014-03-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of GSK-3 inhibition by N-terminal phosphorylation and by the Wnt receptor LRP6.
Elife, 3, 2014
|
|
4ACB
 
 | | CRYSTAL STRUCTURE OF TRANSLATION ELONGATION FACTOR SELB FROM METHANOCOCCUS MARIPALUDIS IN COMPLEX WITH THE GTP ANALOGUE GPPNHP | | Descriptor: | (3ALPHA,5BETA,12ALPHA)-3,12-DIHYDROXYCHOLAN-24-OIC ACID, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-MONOPHOSPHATE, ... | | Authors: | Leibundgut, M, Frick, C, Thanbichler, M, Boeck, A, Ban, N. | | Deposit date: | 2011-12-14 | | Release date: | 2012-11-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.34 Å) | | Cite: | Selenocysteine tRNA-Specific Elongation Factor Selb is a Structural Chimaera of Elongation and Initiation Factors.
Embo J., 24, 2005
|
|
4AC9
 
 | | CRYSTAL STRUCTURE OF TRANSLATION ELONGATION FACTOR SELB FROM METHANOCOCCUS MARIPALUDIS IN COMPLEX WITH GDP | | Descriptor: | (3ALPHA,5BETA,12ALPHA)-3,12-DIHYDROXYCHOLAN-24-OIC ACID, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-MONOPHOSPHATE, ... | | Authors: | Leibundgut, M, Frick, C, Thanbichler, M, Boeck, A, Ban, N. | | Deposit date: | 2011-12-14 | | Release date: | 2012-08-22 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | Selenocysteine tRNA-Specific Elongation Factor Selb is a Structural Chimaera of Elongation and Initiation Factors.
Embo J., 24, 2005
|
|
4ACA
 
 | | CRYSTAL STRUCTURE OF TRANSLATION ELONGATION FACTOR SELB FROM METHANOCOCCUS MARIPALUDIS, APO FORM | | Descriptor: | (3ALPHA,5BETA,12ALPHA)-3,12-DIHYDROXYCHOLAN-24-OIC ACID, GUANOSINE-5'-MONOPHOSPHATE, SULFATE ION, ... | | Authors: | Leibundgut, M, Frick, C, Thanbichler, M, Boeck, A, Ban, N. | | Deposit date: | 2011-12-14 | | Release date: | 2012-11-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Selenocysteine tRNA-Specific Elongation Factor Selb is a Structural Chimaera of Elongation and Initiation Factors.
Embo J., 24, 2005
|
|
4PKS
 
 | | Anthrax toxin lethal factor with bound small molecule inhibitor 11 | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Lethal factor, ... | | Authors: | Maize, K.M, De la Mora, T, Finzel, B.C. | | Deposit date: | 2014-05-15 | | Release date: | 2014-11-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Anthrax toxin lethal factor domain 3 is highly mobile and responsive to ligand binding.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
6U6V
 
 | | Crystal structure of human PD-1H / VISTA | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, V-type immunoglobulin domain-containing suppressor of T-cell activation | | Authors: | Slater, B.T, Han, X, Chen, L, Xiong, Y. | | Deposit date: | 2019-08-30 | | Release date: | 2020-01-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insight into T cell coinhibition by PD-1H (VISTA).
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
