2DLD
 
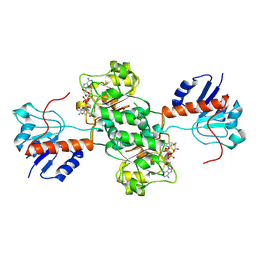 | | D-LACTATE DEHYDROGENASE COMPLEXED WITH NADH AND OXAMATE | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, D-LACTATE DEHYDROGENASE, OXAMIC ACID | | Authors: | Dunn, C.R, Holbrook, J.J. | | Deposit date: | 1995-10-28 | | Release date: | 1996-03-14 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Dehydrogenases Engineering to Correct Substrate Inhibition in a Commercial Dehydrogenase
To be Published
|
|
2HU2
 
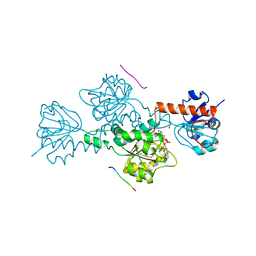 | | CTBP/BARS in ternary complex with NAD(H) and RRTGAPPAL peptide | | Descriptor: | 9-mer peptide from Zinc finger protein 217, C-terminal-binding protein 1, FORMIC ACID, ... | | Authors: | Nardini, M, Bolognesi, M, Quinlan, K.G.R, Verger, A, Francescato, P, Crossley, M. | | Deposit date: | 2006-07-26 | | Release date: | 2006-10-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Specific Recognition of ZNF217 and Other Zinc Finger Proteins at a Surface Groove of C-Terminal Binding Proteins
Mol.Cell.Biol., 26, 2006
|
|
2J6I
 
 | |
2FSS
 
 | |
2GCG
 
 | | Ternary Crystal Structure of Human Glyoxylate Reductase/Hydroxypyruvate Reductase | | Descriptor: | (2R)-2,3-DIHYDROXYPROPANOIC ACID, Glyoxylate reductase/hydroxypyruvate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Booth, M.P.S, Conners, R, Rumsby, G, Brady, R.L. | | Deposit date: | 2006-03-14 | | Release date: | 2006-07-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of substrate specificity in human glyoxylate reductase/hydroxypyruvate reductase
J.Mol.Biol., 360, 2006
|
|
2H1S
 
 | |
2GUG
 
 | | NAD-dependent formate dehydrogenase from Pseudomonas sp.101 in complex with formate | | Descriptor: | DI(HYDROXYETHYL)ETHER, FORMIC ACID, Formate dehydrogenase, ... | | Authors: | Filippova, E.V, Polyakov, K.M, Tikhonova, T.V, Boiko, K.M, Tishkov, V.I, Popov, V.O. | | Deposit date: | 2006-04-30 | | Release date: | 2006-05-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystal structure of the complex of NAD-dependent formate dehydrogenase from
metylotrophic bacterium Pseudomonas sp.101 with formate.
KRISTALLOGRAFIYA, 51, 2006
|
|
2GSD
 
 | | NAD-dependent formate dehydrogenase from bacterium Moraxella sp.C2 in complex with NAD and azide | | Descriptor: | AZIDE ION, NAD-dependent formate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Filippova, E.V, Polyakov, K.M, Tikhonova, T.V, Sadykhov, I.G, Shabalin, I.G, Tishkov, V.I, Popov, V.O. | | Deposit date: | 2006-04-26 | | Release date: | 2006-05-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structures of the apo and holo forms of formate dehydrogenase from the bacterium Moraxella sp. C-1: towards understanding the mechanism of the closure of the interdomain cleft.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
2GO1
 
 | | NAD-dependent formate dehydrogenase from Pseudomonas sp.101 | | Descriptor: | NAD-dependent formate dehydrogenase, SULFATE ION | | Authors: | Filippova, E.V, Polyakov, K.M, Tikhonova, T.V, Stekhanova, T.N, Boiko, K.M, Popov, V.O. | | Deposit date: | 2006-04-12 | | Release date: | 2006-05-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of a new crystal modification of the bacterial NAD-dependent formate dehydrogenase with a resolution of 2.1 A
Crystallography reports, 50, 2005
|
|
2O4C
 
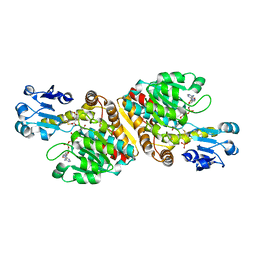 | | Crystal Structure of D-Erythronate-4-phosphate Dehydrogenase Complexed with NAD | | Descriptor: | Erythronate-4-phosphate dehydrogenase, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Ha, J.Y, Lee, J.H, Kim, K.H, Kim, D.J, Lee, H.H, Kim, H.K, Yoon, H.J, Suh, S.W. | | Deposit date: | 2006-12-04 | | Release date: | 2007-02-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of d-Erythronate-4-phosphate Dehydrogenase Complexed with NAD
J.Mol.Biol., 366, 2007
|
|
2OME
 
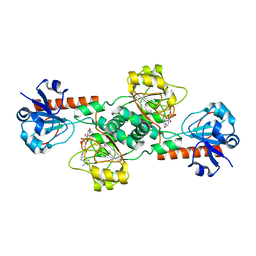 | | Crystal structure of human CTBP2 dehydrogenase complexed with NAD(H) | | Descriptor: | C-terminal-binding protein 2, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Pilka, E.S, Guo, K, Rojkova, A, Debreczeni, J.E, Kavanagh, K.L, von Delft, F, Arrowsmith, C.H, Weigelt, J, Edwards, A, Sundstrom, M, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-01-22 | | Release date: | 2007-02-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of human CTBP2 dehydrogenase complexed with NAD(H)
To be Published
|
|
2NAD
 
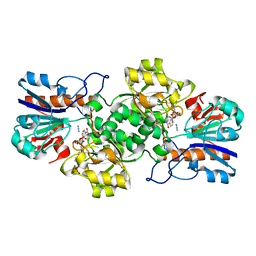 | | HIGH RESOLUTION STRUCTURES OF HOLO AND APO FORMATE DEHYDROGENASE | | Descriptor: | AZIDE ION, NAD-DEPENDENT FORMATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Lamzin, V.S, Dauter, Z, Popov, V.O, Harutyunyan, E.H, Wilson, K.S. | | Deposit date: | 1994-07-06 | | Release date: | 1995-01-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | High resolution structures of holo and apo formate dehydrogenase.
J.Mol.Biol., 236, 1994
|
|
2P9C
 
 | |
2PA3
 
 | |
2P9G
 
 | |
2P9E
 
 | |
2NAC
 
 | | HIGH RESOLUTION STRUCTURES OF HOLO AND APO FORMATE DEHYDROGENASE | | Descriptor: | NAD-DEPENDENT FORMATE DEHYDROGENASE, SULFATE ION | | Authors: | Lamzin, V.S, Dauter, Z, Popov, V.O, Harutyunyan, E.H, Wilson, K.S. | | Deposit date: | 1994-07-06 | | Release date: | 1995-01-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High resolution structures of holo and apo formate dehydrogenase.
J.Mol.Biol., 236, 1994
|
|
2Q50
 
 | | Ensemble refinement of the protein crystal structure of a glyoxylate/hydroxypyruvate reductase from Homo sapiens | | Descriptor: | Glyoxylate reductase/hydroxypyruvate reductase | | Authors: | Levin, E.J, Kondrashov, D.A, Wesenberg, G.E, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2007-05-31 | | Release date: | 2007-06-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Ensemble refinement of protein crystal structures: validation and application.
Structure, 15, 2007
|
|
6IH5
 
 | | Crystal structure of Phosphite Dehydrogenase mutant I151R/P176E from Ralstonia sp. 4506 in complex with non-natural cofactor Nicotinamide Cytosine dinucleotide | | Descriptor: | Phosphite dehydrogenase, [[(2S,3S,4R,5S)-5-(3-aminocarbonylpyridin-1-ium-1-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2S,3S,4R,5S)-5-(4-azanyl-2-oxidanylidene-pyrimidin-1-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Song, X, Feng, Y, Liu, Y, Zhao, Z. | | Deposit date: | 2018-09-28 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.468 Å) | | Cite: | Structural Insights into Phosphite Dehydrogenase Variants Favoring a Non-natural Redox Cofactor
Acs Catalysis, 9, 2019
|
|
6IH4
 
 | |
6IH2
 
 | |
6IH8
 
 | |
6IH3
 
 | | Crystal structure of Phosphite Dehydrogenase from Ralstonia sp. 4506 in complex with non-natural cofactor Nicotinamide Cytosine Dinucleotide | | Descriptor: | Phosphite dehydrogenase, [[(2S,3S,4R,5S)-5-(3-aminocarbonylpyridin-1-ium-1-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2S,3S,4R,5S)-5-(4-azanyl-2-oxidanylidene-pyrimidin-1-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Song, X, Feng, Y, Zhao, Z, Liu, Y. | | Deposit date: | 2018-09-28 | | Release date: | 2019-03-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.942 Å) | | Cite: | Structural Insights into Phosphite Dehydrogenase Variants Favoring a Non-natural Redox Cofactor
Acs Catalysis, 9, 2019
|
|
6IH6
 
 | | Phosphite Dehydrogenase mutant I151R/P176R/M207A from Ralstonia sp. 4506 in complex with non-natural cofactor Nicotinamide Cytosine dinucleotide | | Descriptor: | Phosphite dehydrogenase, [[(2S,3S,4R,5S)-5-(3-aminocarbonylpyridin-1-ium-1-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2S,3S,4R,5S)-5-(4-azanyl-2-oxidanylidene-pyrimidin-1-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Song, X, Feng, Y, Liu, Y, Zhao, Z. | | Deposit date: | 2018-09-28 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.491 Å) | | Cite: | Structural Insights into Phosphite Dehydrogenase Variants Favoring a Non-natural Redox Cofactor
Acs Catalysis, 9, 2019
|
|
6JX1
 
 | | Crystal structure of Formate dehydrogenase mutant V198I/C256I/P260S/E261P/S381N/S383F from Pseudomonas sp. 101 | | Descriptor: | Formate dehydrogenase, GLYCEROL | | Authors: | Feng, Y, Xue, S, Guo, X, Zhao, Z. | | Deposit date: | 2019-04-21 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.233 Å) | | Cite: | Structure-Guided Design of Formate Dehydrogenase for Regeneration of a Non-Natural Redox Cofactor.
Chemistry, 26, 2020
|
|
