8GD2
 
 | | Crystal structure of human cellular retinol binding protein 1 in complex with N-methyl-1-{3-[1-(4-methylphenyl)cyclopentyl]-1,2,4-oxadiazol-5-yl}-N-(2-thienylmethyl)methanamine | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, N-methyl-1-{3-[1-(4-methylphenyl)cyclopentyl]-1,2,4-oxadiazol-5-yl}-N-[(thiophen-2-yl)methyl]methanamine, Retinol-binding protein 1 | | Authors: | Plau, J, Golczak, M. | | Deposit date: | 2023-03-03 | | Release date: | 2023-10-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Discovery of Nonretinoid Inhibitors of CRBP1: Structural and Dynamic Insights for Ligand-Binding Mechanisms.
Acs Chem.Biol., 18, 2023
|
|
8GEV
 
 | | Crystal structure of human cellular retinol binding protein 1 in complex with 1-{[3-(diphenylmethyl)-1,2,4-oxadiazol-5-yl]methyl}-4-(methoxymethyl)piperidine | | Descriptor: | 1-{[3-(diphenylmethyl)-1,2,4-oxadiazol-5-yl]methyl}-4-(methoxymethyl)piperidine, Retinol-binding protein 1 | | Authors: | Plau, J, Golczak, M. | | Deposit date: | 2023-03-07 | | Release date: | 2023-10-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery of Nonretinoid Inhibitors of CRBP1: Structural and Dynamic Insights for Ligand-Binding Mechanisms.
Acs Chem.Biol., 18, 2023
|
|
8GEU
 
 | | Crystal structure of human cellular retinol binding protein 1 in complex with methyl({3-[1-(4-methylphenyl)cyclopentyl]-1,2,4-oxadiazol-5-yl}methyl)[(1-methylpyrazol-4-yl)methyl]amine | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, N-methyl-1-{3-[1-(4-methylphenyl)cyclopentyl]-1,2,4-oxadiazol-5-yl}-N-[(1-methyl-1H-pyrazol-4-yl)methyl]methanamine, Retinol-binding protein 1 | | Authors: | Plau, J, Golczak, M. | | Deposit date: | 2023-03-07 | | Release date: | 2023-10-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Discovery of Nonretinoid Inhibitors of CRBP1: Structural and Dynamic Insights for Ligand-Binding Mechanisms.
Acs Chem.Biol., 18, 2023
|
|
8HTA
 
 | |
5WY9
 
 | |
8W00
 
 | |
8VZZ
 
 | |
8VZY
 
 | |
8W02
 
 | |
8WE3
 
 | | Crystal structure of human FABP4 complexed with C7 | | Descriptor: | 2-[(3-chloranyl-2-phenyl-phenyl)amino]-5-fluoranyl-benzoic acid, Fatty acid-binding protein, adipocyte | | Authors: | Xie, H, Chen, G.F, Xu, Y.C, Li, M.J. | | Deposit date: | 2023-09-16 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structure-based design of potent FABP4 inhibitors with high selectivity against FABP3.
Eur.J.Med.Chem., 264, 2023
|
|
8WKH
 
 | | Crystal structure of group 13 allergen from Blomia tropicalis | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, Fatty acid-binding protein | | Authors: | Zhu, K.L, Gong, Y, Cui, Y.B. | | Deposit date: | 2023-09-27 | | Release date: | 2023-11-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Immunobiological properties and structure analysis of group 13 allergen from Blomia tropicalis and its IgE-mediated cross-reactivity.
Int.J.Biol.Macromol., 254, 2023
|
|
8WDX
 
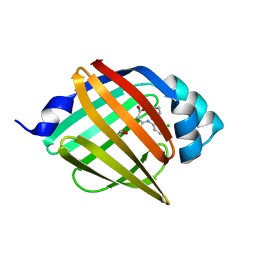 | | Crystal structure of human FABP4 complexed with C3 | | Descriptor: | 1,2-ETHANEDIOL, 2-[(3-chloranyl-2-phenyl-phenyl)amino]-6-methyl-benzoic acid, Fatty acid-binding protein, ... | | Authors: | Xie, H, Chen, G.F, Xu, Y.C. | | Deposit date: | 2023-09-16 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure-based design of potent FABP4 inhibitors with high selectivity against FABP3.
Eur.J.Med.Chem., 264, 2023
|
|
8VZX
 
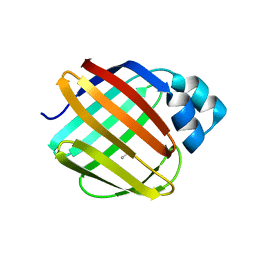 | |
9BCB
 
 | |
7ZLF
 
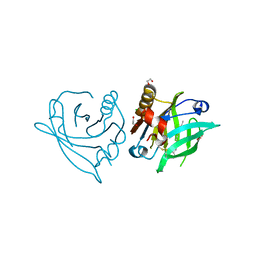 | | Mutant L39Y-L58F of recombinant bovine beta-lactoglobulin in complex with endogenous fatty acid | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactoglobulin, CHLORIDE ION, ... | | Authors: | Loch, J.I, Kurpiewska, K, Bonarek, P. | | Deposit date: | 2022-04-14 | | Release date: | 2023-04-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | beta-Lactoglobulin variants as potential carriers of pramoxine: Comprehensive structural and biophysical studies.
J.Mol.Recognit., 36, 2023
|
|
6VIT
 
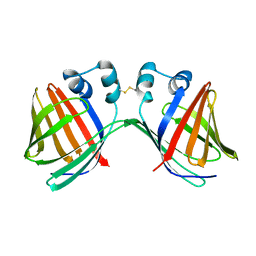 | |
6VID
 
 | |
6VIS
 
 | |
8AEI
 
 | |
8AEH
 
 | |
8AEJ
 
 | |
8AMC
 
 | | Crystal Structure of cat allergen Fel d 4 | | Descriptor: | Allergen Fel d 4, SULFATE ION | | Authors: | Schooltink, L, Sagmeister, T, Gottstein, N, Pavkov-Keller, T, Keller, W. | | Deposit date: | 2022-08-03 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal Structure of cat allergen Fel d 4
To Be Published
|
|
6WNF
 
 | |
6WP1
 
 | |
6WP0
 
 | |
