3SVD
 
 | | Engineered medium-affinity halide-binding protein derived from YFP: bromide complex | | Descriptor: | 1,2-ETHANEDIOL, BROMIDE ION, Green fluorescent protein | | Authors: | Wang, W, Grimley, J.S, Beese, L.S, Hellinga, H.W. | | Deposit date: | 2011-07-12 | | Release date: | 2012-07-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Determination of engineered chloride-binding site structures in fluorescent proteins reveals principles of halide recognition
To be Published
|
|
3TLX
 
 | | Crystal Structure of PF10_0086, adenylate kinase from plasmodium falciparum | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Wernimont, A.K, Loppnau, P, Crombet, L, Weadge, J, Perieteanu, A, Edwards, A.M, Arrowsmith, C.H, Park, H, Bountra, C, Hui, R, Amani, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-08-30 | | Release date: | 2011-10-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal Structure of PF10_0086, adenylate kinase from plasmodium falciparum
TO BE PUBLISHED
|
|
3U2D
 
 | | S. aureus GyrB ATPase domain in complex with small molecule inhibitor | | Descriptor: | 4-bromo-5-methyl-N-[1-(3-nitropyridin-2-yl)piperidin-4-yl]-1H-pyrrole-2-carboxamide, DNA gyrase subunit B, MAGNESIUM ION | | Authors: | Boriack-Sjodin, P.A, Prince, D.B, Eakin, A.E, Sherer, B.A. | | Deposit date: | 2011-10-03 | | Release date: | 2012-01-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Pyrrolamide DNA gyrase inhibitors: fragment-based nuclear magnetic resonance screening to identify antibacterial agents.
Antimicrob.Agents Chemother., 56, 2012
|
|
3U3K
 
 | | Crystal structure of hSULT1A1 bound to PAP and 2-Naphtol | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, Sulfotransferase 1A1, naphthalen-2-ol | | Authors: | Guttman, C, Berger, I, Aharoni, A, Zarivach, R. | | Deposit date: | 2011-10-06 | | Release date: | 2011-11-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | The molecular basis for the broad substrate specificity of human sulfotransferase 1A1.
Plos One, 6, 2011
|
|
3U6J
 
 | | Crystal structure of the VEGFR2 kinase domain in complex with a pyrazolone inhibitor | | Descriptor: | N-{4-[(6,7-dimethoxyquinolin-4-yl)oxy]-3-fluorophenyl}-1,5-dimethyl-3-oxo-2-phenyl-2,3-dihydro-1H-pyrazole-4-carboxamide, Vascular endothelial growth factor receptor 2 | | Authors: | Whittington, D.A, Long, A, Rose, P, Gu, Y, Zhao, H. | | Deposit date: | 2011-10-12 | | Release date: | 2012-02-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure-based design of novel class II c-Met inhibitors: 1. Identification of pyrazolone-based derivatives.
J.Med.Chem., 55, 2012
|
|
3TOL
 
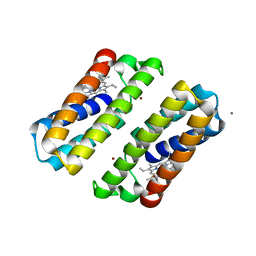 | | Crystal structure of an engineered cytochrome cb562 that forms 1D, Zn-mediated coordination polymers | | Descriptor: | CALCIUM ION, PROTOPORPHYRIN IX CONTAINING FE, Soluble cytochrome b562, ... | | Authors: | Brodin, J.B, Tezcan, F.A. | | Deposit date: | 2011-09-05 | | Release date: | 2012-07-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Metal-directed, chemically tunable assembly of one-, two- and three-dimensional crystalline protein arrays.
Nat Chem, 4, 2012
|
|
3TQ5
 
 | |
3U7U
 
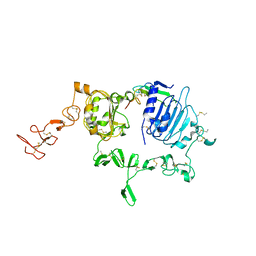 | | Crystal structure of extracellular region of human epidermal growth factor receptor 4 in complex with neuregulin-1 beta | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Neuregulin 1, Receptor tyrosine-protein kinase erbB-4 | | Authors: | Liu, P, Cleveland IV, T.E, Bouyain, S, Longo, P.A, Leahy, D.J. | | Deposit date: | 2011-10-14 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | A single ligand is sufficient to activate EGFR dimers.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3U9Q
 
 | |
3TQW
 
 | | Structure of a ABC transporter, periplasmic substrate-binding protein from Coxiella burnetii | | Descriptor: | METHIONINE, Methionine-binding protein, SULFATE ION | | Authors: | Cheung, J, Franklin, M.C, Rudolph, M, Cassidy, M, Gary, E, Burshteyn, F, Love, J. | | Deposit date: | 2011-09-09 | | Release date: | 2011-09-21 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural genomics for drug design against the pathogen Coxiella burnetii.
Proteins, 83, 2015
|
|
3TRC
 
 | | Structure of the GAF domain from a phosphoenolpyruvate-protein phosphotransferase (ptsP) from Coxiella burnetii | | Descriptor: | PHOSPHATE ION, Phosphoenolpyruvate-protein phosphotransferase, SODIUM ION | | Authors: | Cheung, J, Franklin, M.C, Rudolph, M, Cassidy, M, Gary, E, Burshteyn, F, Love, J. | | Deposit date: | 2011-09-09 | | Release date: | 2011-09-28 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural genomics for drug design against the pathogen Coxiella burnetii.
Proteins, 83, 2015
|
|
3U9Z
 
 | | Crystal structure between actin and a protein construct containing the first beta-thymosin domain of drosophila ciboulot (residues 2-58) with the three mutations N26D/Q27K/D28S | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Renault, L, Husson, C, Carlier, M.F, Didry, D. | | Deposit date: | 2011-10-20 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | How a single residue in individual beta-thymosin/WH2 domains controls their functions in actin assembly
Embo J., 31, 2012
|
|
3TRK
 
 | | Structure of the Chikungunya virus nsP2 protease | | Descriptor: | Nonstructural polyprotein, SODIUM ION | | Authors: | Cheung, J, Franklin, M, Mancia, F, Rudolph, M, Cassidy, M, Gary, E, Burshteyn, F, Love, J. | | Deposit date: | 2011-09-09 | | Release date: | 2011-09-28 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.397 Å) | | Cite: | Structure of the Chikungunya virus nsP2 protease
To be Published
|
|
3UA8
 
 | | Crystal Structure Analysis of a 6-Amino Quinazolinedione Sulfonamide bound to human GluR2 | | Descriptor: | Glutamate receptor 2, N-methyl-1-{3-[(methylsulfonyl)amino]-2,4-dioxo-7-(trifluoromethyl)-1,2,3,4-tetrahydroquinazolin-6-yl}-1H-imidazole-4-carboxamide | | Authors: | Kallen, J. | | Deposit date: | 2011-10-21 | | Release date: | 2012-01-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 6-Amino quinazolinedione sulfonamides as orally active competitive AMPA receptor antagonists.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
3TIH
 
 | |
3TKR
 
 | | Crystal structure of full-length human peroxiredoxin 4 with T118E mutation | | Descriptor: | Peroxiredoxin-4 | | Authors: | Wang, X, Wang, L, Wang, X, Sun, F, Wang, C.-C. | | Deposit date: | 2011-08-28 | | Release date: | 2011-10-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insights into the peroxidase activity and inactivation of human peroxiredoxin 4
Biochem.J., 2011
|
|
3TLC
 
 | |
3TLW
 
 | | The GLIC pentameric Ligand-Gated Ion Channel Loop2-21' oxidized mutant in a locally-closed conformation (LC2 subtype) | | Descriptor: | CHLORIDE ION, DODECYL-BETA-D-MALTOSIDE, Glr4197 protein | | Authors: | Sauguet, L, Nury, H, Corringer, P.J, Delarue, M. | | Deposit date: | 2011-08-30 | | Release date: | 2012-05-16 | | Last modified: | 2012-06-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A locally closed conformation of a bacterial pentameric proton-gated ion channel.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3TN9
 
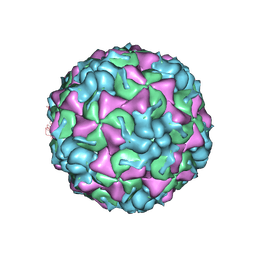 | | X-ray structure of the HRV2 empty capsid (B-particle) | | Descriptor: | Protein VP1, Protein VP2, Protein VP3 | | Authors: | Garriga, D, Pickl-Herk, A, Luque, D, Wruss, J, Caston, J.R, Blaas, D, Verdaguer, N. | | Deposit date: | 2011-09-01 | | Release date: | 2012-03-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Insights into minor group rhinovirus uncoating: the X-ray structure of the HRV2 empty capsid.
Plos Pathog., 8, 2012
|
|
3TTF
 
 | | Crystal structure of E. coli HypF with AMP and carbamoyl phosphate | | Descriptor: | ADENOSINE MONOPHOSPHATE, MAGNESIUM ION, Transcriptional regulatory protein, ... | | Authors: | Petkun, S, Shi, R, Li, Y, Cygler, M. | | Deposit date: | 2011-09-14 | | Release date: | 2011-12-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structure of Hydrogenase Maturation Protein HypF with Reaction Intermediates Shows Two Active Sites.
Structure, 19, 2011
|
|
3TPM
 
 | |
3TTT
 
 | | Structure of F413Y variant of E. coli KatE | | Descriptor: | Catalase HPII, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Loewen, P.C, Jha, V. | | Deposit date: | 2011-09-15 | | Release date: | 2011-10-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Mutation of Phe413 to Tyr in catalase KatE from Escherichia coli leads to side chain damage and main chain cleavage.
Arch.Biochem.Biophys., 525, 2012
|
|
3TQL
 
 | | Structure of the amino acid ABC transporter, periplasmic amino acid-binding protein from Coxiella burnetii. | | Descriptor: | ARGININE, Arginine-binding protein, IMIDAZOLE | | Authors: | Rudolph, M, Cheung, J, Franklin, M.C, Cassidy, M, Gary, E, Burshteyn, F, Love, J. | | Deposit date: | 2011-09-09 | | Release date: | 2011-10-05 | | Last modified: | 2016-01-20 | | Method: | X-RAY DIFFRACTION (1.594 Å) | | Cite: | Structural genomics for drug design against the pathogen Coxiella burnetii.
Proteins, 83, 2015
|
|
3TQR
 
 | | Structure of the phosphoribosylglycinamide formyltransferase (purN) in complex with CHES from Coxiella burnetii | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, CHLORIDE ION, Phosphoribosylglycinamide formyltransferase | | Authors: | Rudolph, M, Cheung, J, Franklin, M.C, Cassidy, M, Gary, E, Burshteyn, F, Love, J. | | Deposit date: | 2011-09-09 | | Release date: | 2011-09-28 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural genomics for drug design against the pathogen Coxiella burnetii.
Proteins, 83, 2015
|
|
3TR2
 
 | | Structure of a orotidine 5'-phosphate decarboxylase (pyrF) from Coxiella burnetii | | Descriptor: | Orotidine 5'-phosphate decarboxylase | | Authors: | Cheung, J, Franklin, M, Rudolph, M, Cassidy, M, Gary, E, Burshteyn, F, Love, J. | | Deposit date: | 2011-09-09 | | Release date: | 2011-09-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Structural genomics for drug design against the pathogen Coxiella burnetii.
Proteins, 83, 2015
|
|
