5LBH
 
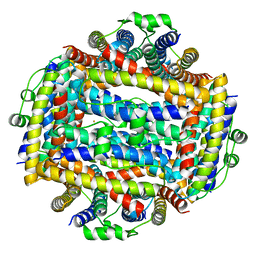 | | Crystal structure of Helicobacter cinaedi CAIP | | Descriptor: | CAIP, FE (III) ION | | Authors: | Zanotti, G, Valesse, F, Codolo, G, De Bernard, M. | | Deposit date: | 2016-06-16 | | Release date: | 2017-04-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.553 Å) | | Cite: | The Helicobacter cinaedi antigen CAIP participates in atherosclerotic inflammation by promoting the differentiation of macrophages in foam cells.
Sci Rep, 7, 2017
|
|
5NVL
 
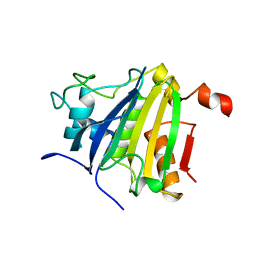 | | Crystal structure of the human 4EHP-GIGYF2 complex | | Descriptor: | Eukaryotic translation initiation factor 4E type 2, GRB10-interacting GYF protein 2 | | Authors: | Peter, D, Valkov, E. | | Deposit date: | 2017-05-04 | | Release date: | 2017-07-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | GIGYF1/2 proteins use auxiliary sequences to selectively bind to 4EHP and repress target mRNA expression.
Genes Dev., 31, 2017
|
|
8REB
 
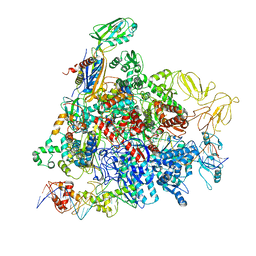 | |
8REA
 
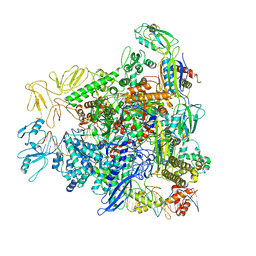 | |
7ZJ1
 
 | | Crystal structure of ADAR1-dsRBD3 dimer | | Descriptor: | Double-stranded RNA-specific adenosine deaminase | | Authors: | Mboukou, A, Barraud, P. | | Deposit date: | 2022-04-08 | | Release date: | 2023-10-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Dimerization of ADAR1 modulates site-specificity of RNA editing
Biorxiv, 2023
|
|
8RE4
 
 | |
8RED
 
 | |
8REE
 
 | |
8REC
 
 | |
8R23
 
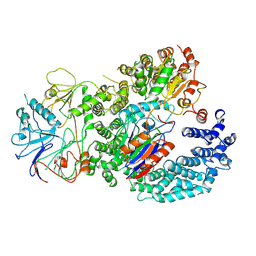 | | INTS9-INTS11-BRAT1 complex | | Descriptor: | BRCA1-associated ATM activator 1, Integrator complex subunit 11, Integrator complex subunit 9, ... | | Authors: | Sabath, K, Qiu, C, Jonas, S. | | Deposit date: | 2023-11-02 | | Release date: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Assembly mechanism of Integrator's RNA cleavage module.
Mol.Cell, 2024
|
|
8R22
 
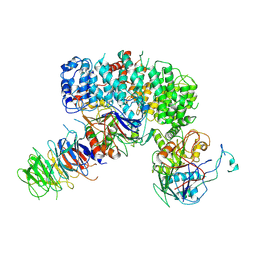 | | INTS9-INTS11-BRAT1-WDR73 complex | | Descriptor: | BRCA1-associated ATM activator 1, Integrator complex subunit 11, Integrator complex subunit 9, ... | | Authors: | Sabath, K, Qiu, C, Jonas, S. | | Deposit date: | 2023-11-02 | | Release date: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Assembly mechanism of Integrator's RNA cleavage module.
Mol.Cell, 2024
|
|
5NVM
 
 | |
2XB2
 
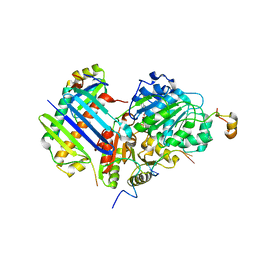 | | Crystal structure of the core Mago-Y14-eIF4AIII-Barentsz-UPF3b assembly shows how the EJC is bridged to the NMD machinery | | Descriptor: | EUKARYOTIC INITIATION FACTOR 4A-III, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Buchwald, G, Ebert, J, Basquin, C, Sauliere, J, Jayachandran, U, Bono, F, Le Hir, H, Conti, E. | | Deposit date: | 2010-04-03 | | Release date: | 2010-05-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Insights Into the Recruitment of the Nmd Machinery from the Crystal Structure of a Core Ejc-Upf3B Complex.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
5NVK
 
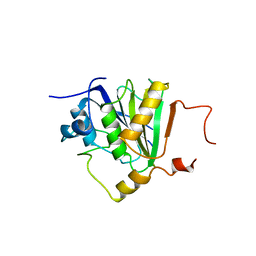 | | Crystal structure of the human 4EHP-GIGYF1 complex | | Descriptor: | Eukaryotic translation initiation factor 4E type 2, GRB10-interacting GYF protein 1 | | Authors: | Peter, D, Valkov, E. | | Deposit date: | 2017-05-04 | | Release date: | 2017-07-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | GIGYF1/2 proteins use auxiliary sequences to selectively bind to 4EHP and repress target mRNA expression.
Genes Dev., 31, 2017
|
|
6EIH
 
 | |
4NZ0
 
 | |
4NLF
 
 | |
4NYZ
 
 | | The EMCV 3Dpol structure with altered motif A conformation at 2.15A resolution | | Descriptor: | GLUTAMINE, GLYCEROL, Genome polyprotein, ... | | Authors: | Vives-adrian, L, Lujan, C, Ferrer-orta, C, Verdaguer, N. | | Deposit date: | 2013-12-11 | | Release date: | 2014-03-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The crystal structure of a cardiovirus RNA-dependent RNA polymerase reveals an unusual conformation of the polymerase active site
J.Virol., 88, 2014
|
|
7XT3
 
 | | Crystal Structure of Hepatitis virus A 2C protein 128-335 aa | | Descriptor: | Genome polyprotein, PHOSPHATE ION | | Authors: | Chen, P, Wojdyla, J.A, Li, Z, Wang, M, Cui, S. | | Deposit date: | 2022-05-16 | | Release date: | 2022-07-27 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Biochemical and structural characterization of hepatitis A virus 2C reveals an unusual ribonuclease activity on single-stranded RNA.
Nucleic Acids Res., 50, 2022
|
|
4K3H
 
 | | Immunoglobulin lambda variable domain L5(L89S) fluorogen activationg protein in complex with malachite green | | Descriptor: | 4-{bis[4-(dimethylamino)phenyl]methyl}phenol, GLYCEROL, Immunoglobulin lambda variable domain L5(L89S), ... | | Authors: | Stanfield, R.L, Szent-Gyorgyi, C, Wilson, I.A. | | Deposit date: | 2013-04-10 | | Release date: | 2013-10-09 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Malachite Green Mediates Homodimerization of Antibody VL Domains to Form a Fluorescent Ternary Complex with Singular Symmetric Interfaces.
J.Mol.Biol., 425, 2013
|
|
5ZW3
 
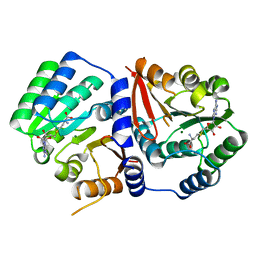 | | Crystal Structure of TrmR from B. subtilis | | Descriptor: | MAGNESIUM ION, Putative O-methyltransferase YrrM, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Kim, J, Ryu, H, Almo, S.C. | | Deposit date: | 2018-05-14 | | Release date: | 2018-07-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Identification of a novel tRNA wobble uridine modifying activity in the biosynthesis of 5-methoxyuridine.
Nucleic Acids Res., 46, 2018
|
|
1ETU
 
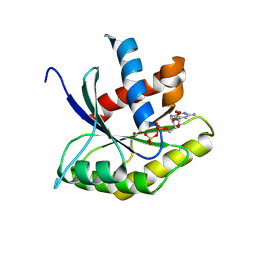 | | STRUCTURAL DETAILS OF THE BINDING OF GUANOSINE DIPHOSPHATE TO ELONGATION FACTOR TU FROM E. COLI AS STUDIED BY X-RAY CRYSTALLOGRAPHY | | Descriptor: | ELONGATION FACTOR TU, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Clark, B.F.C, Lacour, T.F.M, Kjeldgaard, M, Morikawa, K, Nyborg, J, Rubin, R, Thirup, S. | | Deposit date: | 1988-01-15 | | Release date: | 1988-07-16 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural details of the binding of guanosine diphosphate to elongation factor Tu from E. coli as studied by X-ray crystallography.
EMBO J., 4, 1985
|
|
2LYD
 
 | |
7Z2J
 
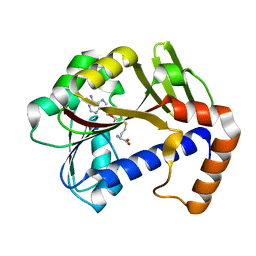 | | White Bream virus N7-Methyltransferase | | Descriptor: | Non-structural protein 1, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Shannon, A, Gauffre, P, Canard, B, Ferron, F. | | Deposit date: | 2022-02-28 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.657 Å) | | Cite: | A second type of N7-guanine RNA cap methyltransferase in an unusual locus of a large RNA virus genome.
Nucleic Acids Res., 50, 2022
|
|
8PDY
 
 | | E. coli RNA polymerase paused at ops site | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Zuber, P.K, Said, N, Hilal, T, Loll, B, Wahl, M.C, Knauer, S.H. | | Deposit date: | 2023-06-13 | | Release date: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Concerted transformation of a hyper-paused transcription complex and its reinforcing protein.
Nat Commun, 15, 2024
|
|
