1QM0
 
 | |
2EU0
 
 | | The NMR ensemble structure of the Itk SH2 domain bound to a phosphopeptide | | Descriptor: | Lymphocyte cytosolic protein 2 phosphopeptide fragment, Tyrosine-protein kinase ITK/TSK | | Authors: | Sundd, M, Pletneva, E.V, Fulton, D.B, Andreotti, A.H. | | Deposit date: | 2005-10-27 | | Release date: | 2006-02-07 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Molecular Details of Itk Activation by Prolyl Isomerization and Phospholigand Binding: The NMR Structure of the Itk SH2 Domain Bound to a Phosphopeptide.
J.Mol.Biol., 357, 2006
|
|
1MPH
 
 | | PLECKSTRIN HOMOLOGY DOMAIN FROM MOUSE BETA-SPECTRIN, NMR, 50 STRUCTURES | | Descriptor: | BETA SPECTRIN | | Authors: | Nilges, M, Macias, M.J, O'Donoghue, S.I, Oschkinat, H. | | Deposit date: | 1997-04-23 | | Release date: | 1997-06-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Automated NOESY interpretation with ambiguous distance restraints: the refined NMR solution structure of the pleckstrin homology domain from beta-spectrin.
J.Mol.Biol., 269, 1997
|
|
1DX0
 
 | | BOVINE PRION PROTEIN RESIDUES 23-230 | | Descriptor: | PRION PROTEIN | | Authors: | Lopez-Garcia, F, Zahn, R, Riek, R, Billeter, M, Wuthrich, K. | | Deposit date: | 1999-12-15 | | Release date: | 2000-07-20 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the Bovine Prion Protein
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1OCA
 
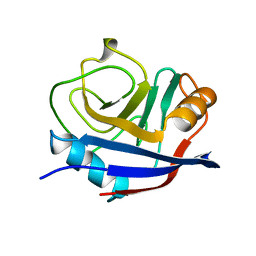 | | HUMAN CYCLOPHILIN A, UNLIGATED, NMR, 20 STRUCTURES | | Descriptor: | CYCLOPHILIN A | | Authors: | Ottiger, M, Zerbe, O, Guntert, P, Wuthrich, K. | | Deposit date: | 1997-07-07 | | Release date: | 1997-11-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The NMR solution conformation of unligated human cyclophilin A.
J.Mol.Biol., 272, 1997
|
|
1Q2N
 
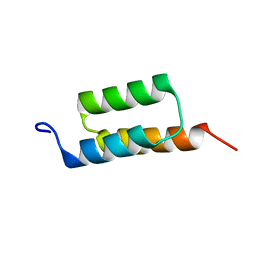 | |
1NYJ
 
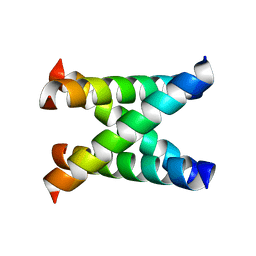 | |
1MIT
 
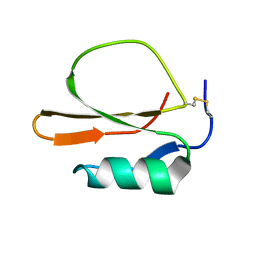 | | RECOMBINANT CUCURBITA MAXIMA TRYPSIN INHIBITOR V (RCMTI-V) (NMR, MINIMIZED AVERAGE STRUCTURE) | | Descriptor: | TRYPSIN INHIBITOR V | | Authors: | Cai, M, Gong, Y, Huang, Y, Liu, J, Prakash, O, Wen, L, Wen, J.J, Huang, J.-K, Krishnamoorthi, R. | | Deposit date: | 1995-10-26 | | Release date: | 1996-04-03 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure and backbone dynamics of recombinant Cucurbita maxima trypsin inhibitor-V determined by NMR spectroscopy.
Biochemistry, 35, 1996
|
|
1DX1
 
 | | BOVINE PRION PROTEIN RESIDUES 23-230 | | Descriptor: | PRION PROTEIN | | Authors: | Lopez Garcia, F, Zahn, R, Riek, R, Billeter, M, Wuthrich, K. | | Deposit date: | 1999-12-15 | | Release date: | 2000-07-20 | | Last modified: | 2015-08-05 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the Bovine Prion Protein
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1DWZ
 
 | | Bovine prion protein fragment 121-230 | | Descriptor: | PRION PROTEIN | | Authors: | Lopez-Garcia, F, Zahn, R, Riek, R, Billeter, M, Wuthrich, K. | | Deposit date: | 1999-12-15 | | Release date: | 2000-07-20 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the Bovine Prion Protein
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1DWY
 
 | | Bovine prion protein fragment 121-230 | | Descriptor: | PRION PROTEIN | | Authors: | Lopez-Garcia, F, Zahn, R, Riek, R, Billeter, M, Wuthrich, K. | | Deposit date: | 1999-12-15 | | Release date: | 2000-07-20 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the Bovine Prion Protein
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1DWL
 
 | | The Ferredoxin-Cytochrome complex using heteronuclear NMR and docking simulation | | Descriptor: | CYTOCHROME C553, FERREDOXIN I, HEME C, ... | | Authors: | Morelli, X, Guerlesquin, F, Czjzek, M, Palma, P.N. | | Deposit date: | 1999-12-08 | | Release date: | 1999-12-10 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR, THEORETICAL MODEL | | Cite: | Heteronuclear NMR and Soft Docking: An Experimental Approach for a Structural Model of the Cytochrome C553-Ferredoxin Complex
Biochemistry, 39, 2000
|
|
1JHB
 
 | |
1HEV
 
 | |
2JX6
 
 | | Structure and membrane interactions of the antibiotic peptide dermadistinctin k by solution and oriented 15N and 31P solid-state NMR spectroscopy | | Descriptor: | Dermadistinctin-K | | Authors: | Mendonca Moraes, C, Verly, R.M, Resende, J.M, Bemquerer, M.P, Pilo-Veloso, D, Valente, A, Almeida, F.C.L, Bechinger, B. | | Deposit date: | 2007-11-08 | | Release date: | 2008-11-11 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Structure and membrane interactions of the antibiotic peptide dermadistinctin K by multidimensional solution and oriented 15N and 31P solid-state NMR spectroscopy
Biophys.J., 96, 2009
|
|
1DAX
 
 | | OXIDISED DESULFOVIBRIO AFRICANUS FERREDOXIN I, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | FERREDOXIN I, IRON/SULFUR CLUSTER | | Authors: | Davy, S.L, Osborne, M.J, Moore, G.R. | | Deposit date: | 1997-12-01 | | Release date: | 1999-01-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Determination of the structure of oxidised Desulfovibrio africanus ferredoxin I by 1H NMR spectroscopy and comparison of its solution structure with its crystal structure.
J.Mol.Biol., 277, 1998
|
|
1EGF
 
 | |
1DFD
 
 | | OXIDISED DESULFOVIBRIO AFRICANUS FERREDOXIN I, NMR, 19 STRUCTURES | | Descriptor: | FERREDOXIN I, IRON/SULFUR CLUSTER | | Authors: | Davy, S.L, Osborne, M.J, Moore, G.R. | | Deposit date: | 1997-12-01 | | Release date: | 1999-01-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Determination of the structure of oxidised Desulfovibrio africanus ferredoxin I by 1H NMR spectroscopy and comparison of its solution structure with its crystal structure.
J.Mol.Biol., 277, 1998
|
|
1MMC
 
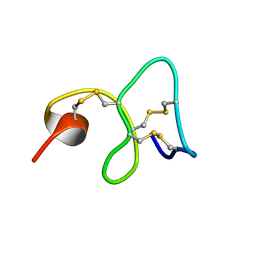 | | 1H NMR STUDY OF THE SOLUTION STRUCTURE OF AC-AMP2 | | Descriptor: | ANTIMICROBIAL PEPTIDE 2 | | Authors: | Martins, J.C, Maes, D, Loris, R, Pepermans, H.A.M, Wyns, L, Willem, R, Verheyden, P. | | Deposit date: | 1995-10-25 | | Release date: | 1996-03-08 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | H NMR study of the solution structure of Ac-AMP2, a sugar binding antimicrobial protein isolated from Amaranthus caudatus.
J.Mol.Biol., 258, 1996
|
|
1RGD
 
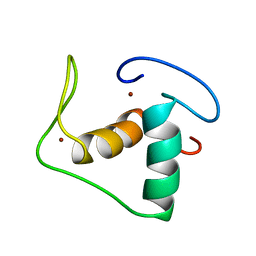 | | STRUCTURE REFINEMENT OF THE GLUCOCORTICOID RECEPTOR-DNA BINDING DOMAIN FROM NMR DATA BY RELAXATION MATRIX CALCULATIONS | | Descriptor: | GLUCOCORTICOID RECEPTOR, ZINC ION | | Authors: | Van Tilborg, M.A.A, Bonvin, A.M.J.J, Hard, K, Davis, A, Maler, B, Boelens, R, Yamamoto, K.R, Kaptein, R. | | Deposit date: | 1995-01-06 | | Release date: | 1995-02-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure refinement of the glucocorticoid receptor-DNA binding domain from NMR data by relaxation matrix calculations.
J.Mol.Biol., 247, 1995
|
|
1UUU
 
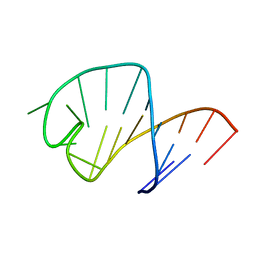 | | STRUCTURE OF AN RNA HAIRPIN LOOP WITH A 5'-CGUUUCG-3' LOOP MOTIF BY HETERONUCLEAR NMR SPECTROSCOPY AND DISTANCE GEOMETRY, 15 STRUCTURES | | Descriptor: | RNA (5'-R(*GP*GP*CP*GP*UP*AP*CP*GP*UP*UP*UP*CP*GP*UP*AP*CP*GP*CP*C)-3') | | Authors: | Sich, C, Ohlenschlager, O, Ramachandran, R, Gorlach, M, Brown, L.R. | | Deposit date: | 1997-08-12 | | Release date: | 1998-02-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of an RNA hairpin loop with a 5'-CGUUUCG-3' loop motif by heteronuclear NMR spectroscopy and distance geometry.
Biochemistry, 36, 1997
|
|
1PBA
 
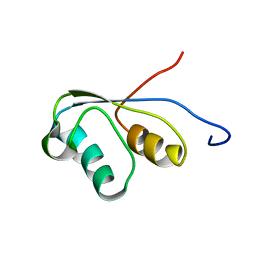 | | THE NMR STRUCTURE OF THE ACTIVATION DOMAIN ISOLATED FROM PORCINE PROCARBOXYPEPTIDASE B | | Descriptor: | PROCARBOXYPEPTIDASE B | | Authors: | Vendrell, J, Wider, G, Billeter, M, Aviles, F.X, Wuthrich, K. | | Deposit date: | 1991-11-18 | | Release date: | 1993-10-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The NMR structure of the activation domain isolated from porcine procarboxypeptidase B.
EMBO J., 10, 1991
|
|
2F40
 
 | |
1HYM
 
 | | HYDROLYZED TRYPSIN INHIBITOR (CMTI-V, MINIMIZED AVERAGE NMR STRUCTURE) | | Descriptor: | HYDROLYZED CUCURBITA MAXIMA TRYPSIN INHIBITOR V | | Authors: | Cai, M, Gong, Y, Prakash, O, Krishnamoorthi, R. | | Deposit date: | 1995-06-12 | | Release date: | 1995-09-15 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Reactive-site hydrolyzed Cucurbita maxima trypsin inhibitor-V: function, thermodynamic stability, and NMR solution structure.
Biochemistry, 34, 1995
|
|
1DAU
 
 | | Analog of dickerson-drew DNA dodecamer with 6'-alpha-methyl carbocyclic thymidines, NMR, minimized average structure | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*(T32)P*(T32)P*CP*GP*CP*G)-3') | | Authors: | Denisov, A, Bekiroglu, S, Maltseva, T, Sandstrom, A, Altmann, K.-H, Egli, M, Chattopadhyaya, J. | | Deposit date: | 1998-01-21 | | Release date: | 1998-05-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution conformation of a carbocyclic analog of the Dickerson-Drew dodecamer: comparison with its own X-ray structure and that of the NMR structure of the native counterpart.
J.Biomol.Struct.Dyn., 16, 1998
|
|
