6NZ3
 
 | |
6NX2
 
 | | Crystal structure of computationally designed protein AAA | | Descriptor: | BROMIDE ION, Design construct AAA | | Authors: | Wei, K.Y, Bick, M.J. | | Deposit date: | 2019-02-07 | | Release date: | 2020-04-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Computational design of closely related proteins that adopt two well-defined but structurally divergent folds.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6NYI
 
 | |
6NYK
 
 | |
1N09
 
 | | A minimal beta-hairpin peptide scaffold for beta-turn display | | Descriptor: | bhpW, disulfide cyclized beta-hairpin peptide | | Authors: | Russell, S.J, Blandl, T, Skelton, N.J, Cochran, A.G. | | Deposit date: | 2002-10-11 | | Release date: | 2003-02-04 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Stability of cyclic beta-hairpins: asymmetric contributions from side chains of a hydrogen-bonded cross-strand residue pair
J.Am.Chem.Soc., 125, 2003
|
|
2MXH
 
 | |
1N0C
 
 | | Stability of cyclic beta-hairpins: Asymmetric contibutions from side chains of hydrogen bonded cross-strand residue pair | | Descriptor: | bhp_HWLV, disulfide cyclized beta-hairpin peptide | | Authors: | Russel, S.J, Blandl, T, Skelton, N.J, Cochran, A.G. | | Deposit date: | 2002-10-11 | | Release date: | 2003-10-21 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Stability of cyclic beta-hairpins: asymmetric contributions from side chains of
a hydrogen-bonded cross-strand residue pair
J.AM.CHEM.SOC., 125, 2003
|
|
2P7C
 
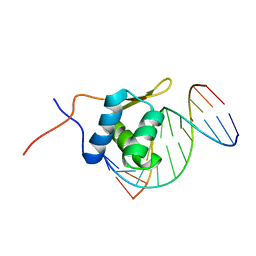 | | Solution structure of the bacillus licheniformis BlaI monomeric form in complex with the blaP half-operator. | | Descriptor: | Penicillinase repressor, Strand 1 of Twelve base-pair DNA, Strand 2 of Twelve base-pair DNA | | Authors: | Boudet, J, Duval, V, Van Melckebeke, H, Blackledge, M, Amoroso, A, Joris, B, Simorre, J.-P. | | Deposit date: | 2007-03-20 | | Release date: | 2007-06-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Conformational and thermodynamic changes of the repressor/DNA operator complex upon monomerization shed new light on regulation mechanisms of bacterial resistance against beta-lactam antibiotics.
Nucleic Acids Res., 35, 2007
|
|
2XXD
 
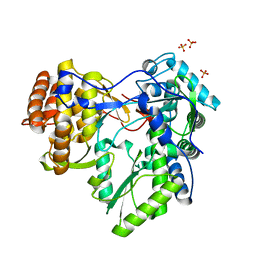 | | HCV-JFH1 NS5B polymerase structure at 1.9 angstrom | | Descriptor: | PHOSPHATE ION, RNA-DIRECTED RNA POLYMERASE | | Authors: | Caillet-Saguy, C, Bressanelli, S. | | Deposit date: | 2010-11-10 | | Release date: | 2011-01-12 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.881 Å) | | Cite: | A Comprehensive Structure-Function Comparison of Hepatitis C Virus Strains Jfh1 and J6 Polymerases Reveals a Key Residue Stimulating Replication in Cell Culture Across Genotypes.
J.Virol., 85, 2011
|
|
2X6P
 
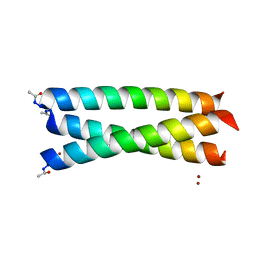 | | Crystal Structure of Coil Ser L19C | | Descriptor: | COIL SER L19C, ZINC ION | | Authors: | Chakraborty, S, Touw, D.S, Peacock, A.F.A, Stuckey, J.A, Pecoraro, V.L. | | Deposit date: | 2010-02-18 | | Release date: | 2010-09-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural Comparisons of Apo- and Metalated Three-Stranded Coiled Coils Clarify Metal Binding Determinants in Thiolate Containing Designed Peptides.
J.Am.Chem.Soc., 132, 2010
|
|
2XYM
 
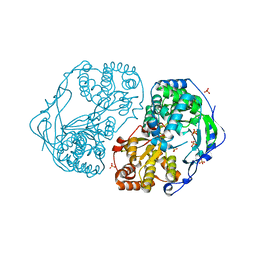 | | HCV-JFH1 NS5B T385A mutant | | Descriptor: | PHOSPHATE ION, RNA-DIRECTED RNA POLYMERASE | | Authors: | Simister, P.C, Caillet-Saguy, C, Bressanelli, S. | | Deposit date: | 2010-11-18 | | Release date: | 2011-01-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.774 Å) | | Cite: | A Comprehensive Structure-Function Comparison of Hepatitis C Virus Strains Jfh1 and J6 Polymerases Reveals a Key Residue Stimulating Replication in Cell Culture Across Genotypes.
J.Virol., 85, 2011
|
|
8WAS
 
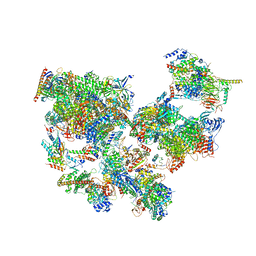 | | Structure of transcribing complex 9 (TC9), the initially transcribing complex with Pol II positioned 9nt downstream of TSS. | | Descriptor: | Alpha-amanitin, CDK-activating kinase assembly factor MAT1, DNA-directed RNA polymerase II subunit E, ... | | Authors: | Chen, X, Liu, W, Wang, Q, Wang, X, Ren, Y, Qu, X, Li, W, Xu, Y. | | Deposit date: | 2023-09-08 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (6.13 Å) | | Cite: | Structural visualization of transcription initiation in action.
Science, 382, 2023
|
|
8WAN
 
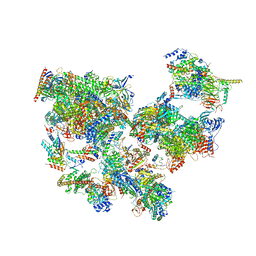 | | Structure of transcribing complex 4 (TC4), the initially transcribing complex with Pol II positioned 4nt downstream of TSS. | | Descriptor: | Alpha-amanitin, CDK-activating kinase assembly factor MAT1, DNA-directed RNA polymerase II subunit E, ... | | Authors: | Chen, X, Liu, W, Wang, Q, Wang, X, Ren, Y, Qu, X, Li, W, Xu, Y. | | Deposit date: | 2023-09-07 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (6.07 Å) | | Cite: | Structural visualization of transcription initiation in action.
Science, 382, 2023
|
|
8WAP
 
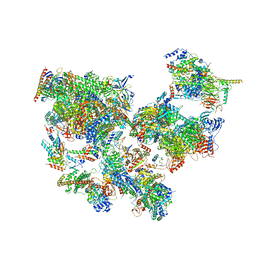 | | Structure of transcribing complex 6 (TC6), the initially transcribing complex with Pol II positioned 6nt downstream of TSS. | | Descriptor: | Alpha-amanitin, CDK-activating kinase assembly factor MAT1, DNA-directed RNA polymerase II subunit E, ... | | Authors: | Chen, X, Liu, W, Wang, Q, Wang, X, Ren, Y, Qu, X, Li, W, Xu, Y. | | Deposit date: | 2023-09-08 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (5.85 Å) | | Cite: | Structural visualization of transcription initiation in action.
Science, 382, 2023
|
|
8WAL
 
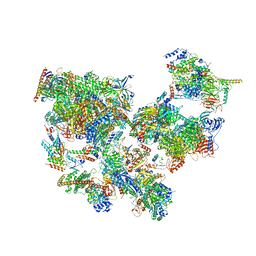 | | Structure of transcribing complex 3 (TC3), the initially transcribing complex with Pol II positioned 3nt downstream of TSS. | | Descriptor: | Alpha-amanitin, CDK-activating kinase assembly factor MAT1, DNA-directed RNA polymerase II subunit E, ... | | Authors: | Chen, X, Liu, W, Wang, Q, Wang, X, Ren, Y, Qu, X, Li, W, Xu, Y. | | Deposit date: | 2023-09-07 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (8.52 Å) | | Cite: | Structural visualization of transcription initiation in action.
Science, 382, 2023
|
|
8WAO
 
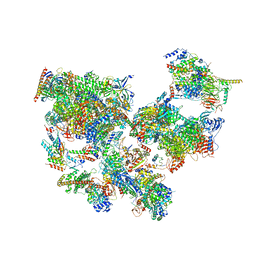 | | Structure of transcribing complex 5 (TC5), the initially transcribing complex with Pol II positioned 5nt downstream of TSS. | | Descriptor: | Alpha-amanitin, CDK-activating kinase assembly factor MAT1, DNA-directed RNA polymerase II subunit E, ... | | Authors: | Chen, X, Liu, W, Wang, Q, Wang, X, Ren, Y, Qu, X, Li, W, Xu, Y. | | Deposit date: | 2023-09-07 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (6.4 Å) | | Cite: | Structural visualization of transcription initiation in action.
Science, 382, 2023
|
|
8WAQ
 
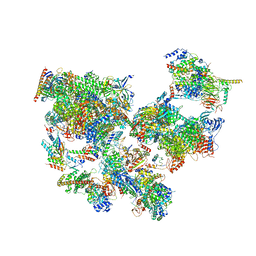 | | Structure of transcribing complex 7 (TC7), the initially transcribing complex with Pol II positioned 7nt downstream of TSS. | | Descriptor: | Alpha-amanitin, CDK-activating kinase assembly factor MAT1, DNA-directed RNA polymerase II subunit E, ... | | Authors: | Chen, X, Liu, W, Wang, Q, Wang, X, Ren, Y, Qu, X, Li, W, Xu, Y. | | Deposit date: | 2023-09-08 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (6.29 Å) | | Cite: | Structural visualization of transcription initiation in action.
Science, 382, 2023
|
|
8WAK
 
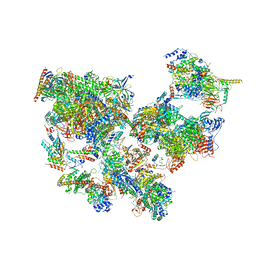 | | Structure of transcribing complex 2 (TC2), the initially transcribing complex with Pol II positioned 2nt downstream of TSS. | | Descriptor: | Alpha-amanitin, CDK-activating kinase assembly factor MAT1, DNA-directed RNA polymerase II subunit E, ... | | Authors: | Chen, X, Liu, W, Wang, Q, Wang, X, Ren, Y, Qu, X, Li, W, Xu, Y. | | Deposit date: | 2023-09-07 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (5.47 Å) | | Cite: | Structural visualization of transcription initiation in action.
Science, 382, 2023
|
|
8WAR
 
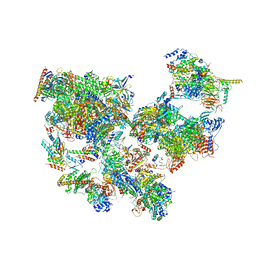 | | Structure of transcribing complex 8 (TC8), the initially transcribing complex with Pol II positioned 8nt downstream of TSS. | | Descriptor: | Alpha-amanitin, CDK-activating kinase assembly factor MAT1, DNA-directed RNA polymerase II subunit E, ... | | Authors: | Chen, X, Liu, W, Wang, Q, Wang, X, Ren, Y, Qu, X, Li, W, Xu, Y. | | Deposit date: | 2023-09-08 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (7.2 Å) | | Cite: | Structural visualization of transcription initiation in action.
Science, 382, 2023
|
|
8A3G
 
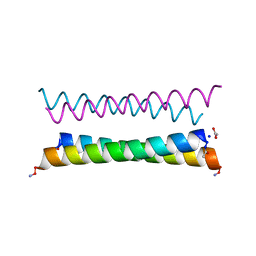 | | X-ray crystal structure of a de novo designed antiparallel coiled-coil homotetramer with 4 heptad repeats, apCC-Tet* | | Descriptor: | ACETATE ION, SODIUM ION, apCC-Tet* | | Authors: | Naudin, E.A, Mylemans, B, Albanese, K.I, Woolfson, D.N. | | Deposit date: | 2022-06-08 | | Release date: | 2022-10-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (0.96 Å) | | Cite: | From peptides to proteins: coiled-coil tetramers to single-chain 4-helix bundles.
Chem Sci, 13, 2022
|
|
8A3J
 
 | | X-ray crystal structure of a de novo designed antiparallel coiled-coil heterotetramer with 3 heptad repeats, apCC-Tet*3-A2B2 | | Descriptor: | apCC-Tet*3-A, apCC-Tet*3-B | | Authors: | Naudin, E.A, Mylemans, B, Albanese, K.I, Woolfson, D.N. | | Deposit date: | 2022-06-08 | | Release date: | 2022-10-05 | | Last modified: | 2022-11-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | From peptides to proteins: coiled-coil tetramers to single-chain 4-helix bundles.
Chem Sci, 13, 2022
|
|
8A3I
 
 | | X-ray crystal structure of a de novo designed antiparallel coiled-coil homotetramer with 3 heptad repeats, apCC-Tet*3 | | Descriptor: | apCC-Tet*3 | | Authors: | Naudin, E.A, Mylemans, B, Albanese, K.I, Woolfson, D.N. | | Deposit date: | 2022-06-08 | | Release date: | 2022-10-05 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | From peptides to proteins: coiled-coil tetramers to single-chain 4-helix bundles.
Chem Sci, 13, 2022
|
|
8A3K
 
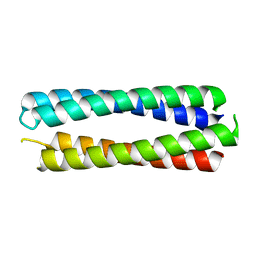 | | X-ray crystal structure of a de novo designed single-chain antiparallel 4-helix coiled-coil bundle, sc-apCC-4 | | Descriptor: | sc-apCC-4 | | Authors: | Albanese, K.I, Mylemans, B, Naudin, E.A, Woolfson, D.N. | | Deposit date: | 2022-06-08 | | Release date: | 2022-10-05 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | From peptides to proteins: coiled-coil tetramers to single-chain 4-helix bundles.
Chem Sci, 13, 2022
|
|
8BCS
 
 | | X-ray crystal structure of a de novo designed helix-loop-helix homodimer in an anti arrangement, CC-HP1.0 | | Descriptor: | ACETATE ION, CC-HP1.0 | | Authors: | Edgell, C.L, Mylemans, B, Naudin, E.A, Smith, A.J, Savery, N.J, Woolfson, D.N. | | Deposit date: | 2022-10-17 | | Release date: | 2023-06-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Design and Selection of Heterodimerizing Helical Hairpins for Synthetic Biology.
Acs Synth Biol, 12, 2023
|
|
8GAC
 
 | | Crystal structure of a high affinity CTLA-4 binder | | Descriptor: | 1,2-ETHANEDIOL, CTLA-4 binder | | Authors: | Yang, W, Almo, S.C, Baker, D, Ghosh, A. | | Deposit date: | 2023-02-22 | | Release date: | 2024-08-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Design of High Affinity Binders to Convex Protein Target Sites.
Biorxiv, 2024
|
|
