4UC8
 
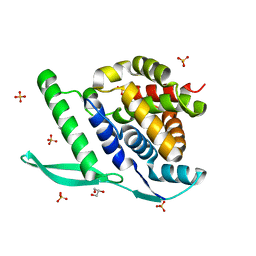 | | N-terminal globular domain of the RSV Nucleoprotein in complex with C- terminal phenylalanine of the Phosphoprotein | | Descriptor: | GLYCEROL, NUCLEOPROTEIN, PHENYLALANINE, ... | | Authors: | Ouizougun-Oubari, M, Pereira, N, Tarus, B, Galloux, M, Tortorici, M.-A, Hoos, S, Baron, B, England, P, Bontems, F, Rey, F.A, Eleouet, J.-F, Sizun, C, Slama-Schwok, A, Duquerroy, S. | | Deposit date: | 2014-12-03 | | Release date: | 2015-08-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Druggable Pocket at the Nucleocapsid/Phosphoprotein Interaction Site of the Human Respiratory Syncytial Virus.
J.Virol., 89, 2015
|
|
7SOA
 
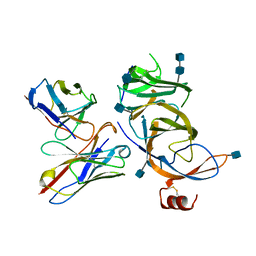 | |
4UC9
 
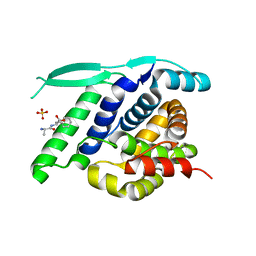 | | N-terminal globular domain of the RSV Nucleoprotein in complex with C- terminal dipeptide of the Phosphoprotein | | Descriptor: | ASPARTIC ACID, NUCLEOPROTEIN, PHENYLALANINE, ... | | Authors: | Ouizougun-Oubari, M, Pereira, N, Tarus, B, Galloux, M, Tortorici, M.-A, Hoos, S, Baron, B, England, P, Bontems, F, Rey, F.A, Eleouet, J.-F, Sizun, C, Slama-Schwok, A, Duquerroy, S. | | Deposit date: | 2014-12-03 | | Release date: | 2015-08-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A Druggable Pocket at the Nucleocapsid/Phosphoprotein Interaction Site of the Human Respiratory Syncytial Virus.
J.Virol., 89, 2015
|
|
7SO9
 
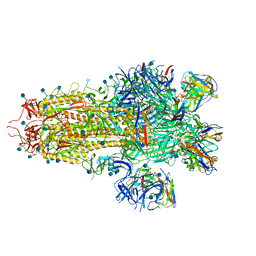 | |
7SOE
 
 | |
4UE6
 
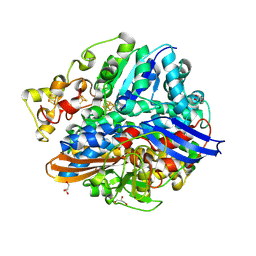 | | Structure of methylene blue-treated anaerobically purified D. fructosovorans NiFe-hydrogenase | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE3-S4 CLUSTER, GLYCEROL, ... | | Authors: | Volbeda, A, Martin, L, Liebgott, P.-P, Fontecilla-Camps, J.C. | | Deposit date: | 2014-12-16 | | Release date: | 2015-03-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | [Nife]-Hydrogenases Revisited: Nickel-Carboxamido Bond Formation in a Variant with Accrued O2-Tolerance and a Tentative Re-Interpretation of Ni-Si States.
Metallomics, 7, 2015
|
|
7SOD
 
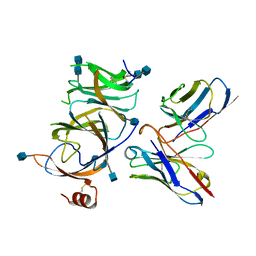 | |
4UDI
 
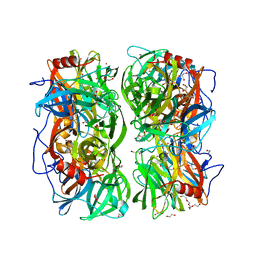 | | Crystal structure of b-1,4-mannopyranosyl-chitobiose phosphorylase at 1.85 Angstrom from unknown human gut bacteria (Uhgb_MP) | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Ladeveze, S, Cioci, G, Potocki-Veronese, G, Tranier, S, Mourey, L. | | Deposit date: | 2014-12-10 | | Release date: | 2015-05-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Bases for N-Glycan Processing by Mannoside Phosphorylase.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
7SOC
 
 | |
4UG3
 
 | | B. subtilis GpsB N-terminal Domain | | Descriptor: | CELL CYCLE PROTEIN GPSB | | Authors: | Rismondo, J, Cleverley, R.M, Lane, H.V, Grohennig, S, Steglich, A, Moller, L, Krishna Mannala, G, Hain, T, Lewis, R.J, Halbedel, S. | | Deposit date: | 2015-03-21 | | Release date: | 2015-11-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the Bacterial Cell Division Determinant Gpsb and its Interaction with Penicillin Binding Proteins.
Mol.Microbiol., 99, 2016
|
|
4UGD
 
 | | Structure of Bacillus subtilis Nitric Oxide Synthase in complex with 6-((((2S)-1-amino-4-((6-amino-4-methylpyridin-2-yl)methoxy)butan-2-yl) oxy)methyl)-4-methylpyridin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6-[({(2S)-1-amino-4-[(6-amino-4-methylpyridin-2-yl)methoxy]butan-2-yl}oxy)methyl]-4-methylpyridin-2-amine, CHLORIDE ION, ... | | Authors: | Holden, J.K, Poulos, T.L. | | Deposit date: | 2015-03-22 | | Release date: | 2015-06-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Inhibitor Bound Crystal Structures of Bacterial Nitric Oxide Synthase.
Biochemistry, 54, 2015
|
|
7SOF
 
 | |
4UF0
 
 | | Crystal structure of JmjC domain of human histone demethylase UTY in complex with epitherapuetic compound 2-(((2-((2-(dimethylamino)ethyl) (ethyl)amino)-2-oxoethyl)amino)methyl)isonicotinic acid. | | Descriptor: | 1,2-ETHANEDIOL, 2-{[(2-{[(E)-2-(dimethylamino)ethenyl](ethyl)amino}-2-oxoethyl)amino]methyl}pyridine-4-carboxylic acid, FE (II) ION, ... | | Authors: | Srikannathasan, V, Johansson, C, Gileadi, C, Tobias, K, Kopec, J, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A, Oppermann, U. | | Deposit date: | 2014-12-22 | | Release date: | 2015-01-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural Analysis of Human Kdm5B Guides Histone Demethylase Inhibitor Development.
Nat.Chem.Biol., 12, 2016
|
|
4UF5
 
 | | Crystal structure of UCH-L5 in complex with inhibitory fragment of INO80G | | Descriptor: | NUCLEAR FACTOR RELATED TO KAPPA-B-BINDING PROTEIN, UBIQUITIN CARBOXYL-TERMINAL HYDROLASE ISOZYME L5 | | Authors: | Sahtoe, D.D, Van Dijk, W.J, El Oualid, F, Ekkebus, R, Ovaa, H, Sixma, T.K. | | Deposit date: | 2014-12-23 | | Release date: | 2015-03-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Mechanism of Uch-L5 Activation and Inhibition by Deubad Domains in Rpn13 and Ino80G.
Mol.Cell, 57, 2015
|
|
4UG8
 
 | | Structure of Bacillus subtilis Nitric Oxide Synthase in complex with 6-(5-((3R,4R)-4-((6-azanyl-4-methyl-pyridin-2-yl)methyl)pyrrolidin-3- yl)oxypentyl)-4-methyl-pyridin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6-[5-({(3R,4R)-4-[(6-amino-4-methylpyridin-2-yl)methyl]pyrrolidin-3-yl}oxy)pentyl]-4-methylpyridin-2-amine, CHLORIDE ION, ... | | Authors: | Holden, J.K, Poulos, T.L. | | Deposit date: | 2015-03-22 | | Release date: | 2015-06-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.888 Å) | | Cite: | Inhibitor Bound Crystal Structures of Bacterial Nitric Oxide Synthase.
Biochemistry, 54, 2015
|
|
4UGL
 
 | | Structure of Bacillus subtilis Nitric Oxide Synthase in complex with N1-(3-(2-(6-Amino-4-methylpyridin-2-yl)ethyl)-5-fluorophenyl)-N1- cyclopropyl-N2-methylethane-1,2-diamine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, CHLORIDE ION, GLYCEROL, ... | | Authors: | Holden, J.K, Poulos, T.L. | | Deposit date: | 2015-03-22 | | Release date: | 2015-06-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Inhibitor Bound Crystal Structures of Bacterial Nitric Oxide Synthase.
Biochemistry, 54, 2015
|
|
4UGC
 
 | | Structure of Bacillus subtilis Nitric Oxide Synthase in complex with 6,6'-(((2S)-3-aminopropane-1,2-diyl)bis(oxymethanediyl))bis(4- methylpyridin-2-amine) | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6,6'-{[(2S)-3-aminopropane-1,2-diyl]bis(oxymethanediyl)}bis(4-methylpyridin-2-amine), CHLORIDE ION, ... | | Authors: | Holden, J.K, Poulos, T.L. | | Deposit date: | 2015-03-22 | | Release date: | 2015-06-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Inhibitor Bound Crystal Structures of Bacterial Nitric Oxide Synthase.
Biochemistry, 54, 2015
|
|
4UGE
 
 | | Structure of Bacillus subtilis Nitric Oxide Synthase in complex with 4-methyl-6-((3-(piperidin-4-ylmethoxy)phenoxy)methyl)pyridin-2-amine | | Descriptor: | 4-methyl-6-{[3-(piperidin-4-ylmethoxy)phenoxy]methyl}pyridin-2-amine, 5,6,7,8-TETRAHYDROBIOPTERIN, CHLORIDE ION, ... | | Authors: | Holden, J.K, Poulos, T.L. | | Deposit date: | 2015-03-22 | | Release date: | 2015-06-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Inhibitor Bound Crystal Structures of Bacterial Nitric Oxide Synthase.
Biochemistry, 54, 2015
|
|
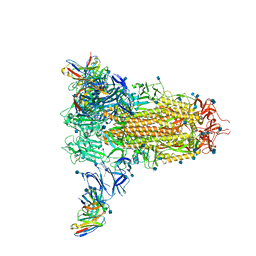
7SN3
 
 | | Structure of human SARS-CoV-2 spike glycoprotein trimer bound by neutralizing antibody C1C-A3 Fab (variable region) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pan, J, Abraham, J, Shankar, S. | | Deposit date: | 2021-10-27 | | Release date: | 2021-12-08 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for continued antibody evasion by the SARS-CoV-2 receptor binding domain.
Science, 375, 2022
|
|
4UGN
 
 | | Structure of Bacillus subtilis Nitric Oxide Synthase in complex with (S)-N-(3-(((Pyrrolidin-2-ylmethyl)amino)methyl)phenyl)thiophene-2- carboximidamide | | Descriptor: | (S)-N-(3-(((Pyrrolidin-2-ylmethyl)amino)methyl)phenyl)thiophene-2-carboximidamide, 5,6,7,8-TETRAHYDROBIOPTERIN, CHLORIDE ION, ... | | Authors: | Holden, J.K, Poulos, T.L. | | Deposit date: | 2015-03-22 | | Release date: | 2015-06-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Inhibitor Bound Crystal Structures of Bacterial Nitric Oxide Synthase.
Biochemistry, 54, 2015
|
|
4UC5
 
 | | Neisseria Meningitidis DAH7PS-Phenylalanine regulated | | Descriptor: | DI(HYDROXYETHYL)ETHER, MANGANESE (II) ION, PHENYLALANINE, ... | | Authors: | Heyes, L.C, Lang, E.J.M, Parker, E.J. | | Deposit date: | 2014-12-03 | | Release date: | 2015-11-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Calculated Pka Variations Expose Dynamic Allosteric Communication Networks.
J.Am.Chem.Soc., 138, 2016
|
|
4UPK
 
 | |
4UGT
 
 | | Structure of Bacillus subtilis Nitric Oxide Synthase in complex with N-(3-((pyrrolidin-3-yloxy)methyl)phenyl)thiophene-2-carboximidamide | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, CHLORIDE ION, GLYCEROL, ... | | Authors: | Holden, J.K, Poulos, T.L. | | Deposit date: | 2015-03-22 | | Release date: | 2015-06-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Inhibitor Bound Crystal Structures of Bacterial Nitric Oxide Synthase.
Biochemistry, 54, 2015
|
|
4UD1
 
 | | Structure of the N Terminal domain of the MERS CoV nucleocapsid | | Descriptor: | AMMONIUM ION, GLYCEROL, IMIDAZOLE, ... | | Authors: | Papageorgiou, N, Lichiere, J, Ferron, F, Canard, B, Coutard, B. | | Deposit date: | 2014-12-05 | | Release date: | 2015-12-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structural Characterization of the N-Terminal Part of the Mers-Cov Nucleocapsid by X-Ray Diffraction and Small-Angle X-Ray Scattering
Acta Crystallogr.,Sect.D, 72, 2016
|
|
4UDT
 
 | | T cell receptor (TRAV22,TRBV7-9) structure | | Descriptor: | GLYCEROL, PROTEIN TRBV7-9, T-CELL RECEPTOR BETA-2 CHAIN C REGION, ... | | Authors: | Rodstrom, K.E.J, Regenthal, P, Lindkvist-Petersson, K. | | Deposit date: | 2014-12-11 | | Release date: | 2015-06-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure of Staphylococcal Enterotoxin E in Complex with Tcr Defines the Role of Tcr Loop Positioning in Superantigen Recognition.
Plos One, 10, 2015
|
|
