7RFP
 
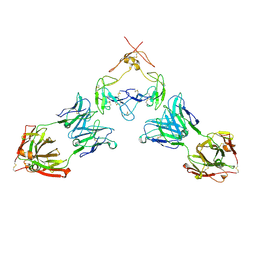 | | Mouse GITR (mGITR) with DTA-1 Fab fragment | | Descriptor: | DTA-1 (heavy chain), DTA-1 (light chain), Tumor necrosis factor receptor superfamily member 18,Enhanced green fluorescent protein | | Authors: | Meyerson, J.R, He, C. | | Deposit date: | 2021-07-14 | | Release date: | 2022-03-09 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Therapeutic antibody activation of the glucocorticoid-induced TNF receptor by a clustering mechanism.
Sci Adv, 8, 2022
|
|
8BSJ
 
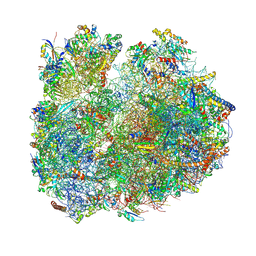 | | Giardia Ribosome in PRE-T Classical State (C) | | Descriptor: | 40S ribosomal protein S21, 40S ribosomal protein S25, 40S ribosomal protein S26, ... | | Authors: | Majumdar, S, Emmerich, A.G, Sanyal, S. | | Deposit date: | 2022-11-25 | | Release date: | 2023-03-22 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (6.49 Å) | | Cite: | Insights into translocation mechanism and ribosome evolution from cryo-EM structures of translocation intermediates of Giardia intestinalis.
Nucleic Acids Res., 51, 2023
|
|
2ECK
 
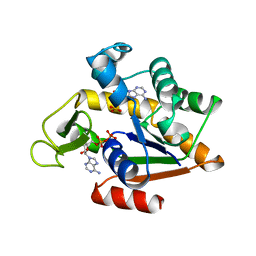 | | STRUCTURE OF PHOSPHOTRANSFERASE | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, ADENYLATE KINASE | | Authors: | Berry, M.B, Bilderback, T, Glaser, M, Phillips Jr, G.N. | | Deposit date: | 1996-12-16 | | Release date: | 1997-03-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of ADP/AMP complex of Escherichia coli adenylate kinase.
Proteins, 62, 2006
|
|
8BTD
 
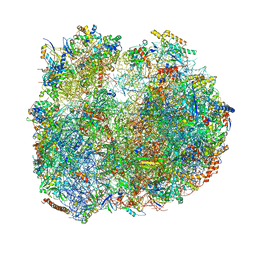 | | Giardia Ribosome in PRE-T Hybrid State (D1) | | Descriptor: | 5.8S rRNA, 5S rRNA, Large Subunit rRNA, ... | | Authors: | Majumdar, S, Emmerich, A.G, Sanyal, S. | | Deposit date: | 2022-11-28 | | Release date: | 2023-03-22 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Insights into translocation mechanism and ribosome evolution from cryo-EM structures of translocation intermediates of Giardia intestinalis.
Nucleic Acids Res., 51, 2023
|
|
8BQX
 
 | | Yeast 80S ribosome in complex with Map1 (conformation 2) | | Descriptor: | 18S rRNA, 25S rRNA, 40S ribosomal protein S0-A, ... | | Authors: | Knorr, A.G, Mackens-Kiani, T, Musial, J, Berninghausen, O, Becker, T, Beatrix, B, Beckmann, R. | | Deposit date: | 2022-11-21 | | Release date: | 2023-03-22 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | The dynamic architecture of Map1- and NatB-ribosome complexes coordinates the sequential modifications of nascent polypeptide chains.
Plos Biol., 21, 2023
|
|
8BQD
 
 | | Yeast 80S ribosome in complex with Map1 (conformation 1) | | Descriptor: | 18S rRNA, 25S rRNA, 40S ribosomal protein S0-A, ... | | Authors: | Knorr, A.G, Mackens-Kiani, T, Musial, J, Berninghausen, O, Becker, T, Beatrix, B, Beckmann, R. | | Deposit date: | 2022-11-21 | | Release date: | 2023-03-22 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | The dynamic architecture of Map1- and NatB-ribosome complexes coordinates the sequential modifications of nascent polypeptide chains.
Plos Biol., 21, 2023
|
|
8BTR
 
 | | Giardia Ribosome in PRE-T Hybrid State (D2) | | Descriptor: | 5.8S rRNA, 5S rRNA, Large Subunit rRNA, ... | | Authors: | Majumdar, S, Emmerich, A.G, Sanyal, S. | | Deposit date: | 2022-11-29 | | Release date: | 2023-03-22 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Insights into translocation mechanism and ribosome evolution from cryo-EM structures of translocation intermediates of Giardia intestinalis.
Nucleic Acids Res., 51, 2023
|
|
8BR8
 
 | | Giardia ribosome in POST-T state (A1) | | Descriptor: | 40S ribosomal protein S21, 40S ribosomal protein S25, 40S ribosomal protein S26, ... | | Authors: | Majumdar, S, Emmerich, A.G, Sanyal, S. | | Deposit date: | 2022-11-22 | | Release date: | 2023-03-15 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Insights into translocation mechanism and ribosome evolution from cryo-EM structures of translocation intermediates of Giardia intestinalis.
Nucleic Acids Res., 51, 2023
|
|
8BRM
 
 | | Giardia ribosome in POST-T state, no E-site tRNA (A6) | | Descriptor: | 5.8S rRNA, 5S rRNA, Large Subunit rRNA, ... | | Authors: | Majumdar, S, Emmerich, A.G, Sanyal, S. | | Deposit date: | 2022-11-23 | | Release date: | 2023-03-15 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Insights into translocation mechanism and ribosome evolution from cryo-EM structures of translocation intermediates of Giardia intestinalis.
Nucleic Acids Res., 51, 2023
|
|
2EPG
 
 | |
7MHT
 
 | | CYTOSINE-SPECIFIC METHYLTRANSFERASE HHAI/DNA COMPLEX | | Descriptor: | 5'-D(P*CP*CP*AP*TP*GP*AP*GP*CP*TP*GP*AP*C)-3', 5'-D(P*GP*TP*CP*AP*GP*CP*GP*CP*AP*TP*GP*G)-3', CYTOSINE-SPECIFIC METHYLTRANSFERASE HHAI, ... | | Authors: | O'Gara, M, Horton, J.R, Roberts, R.J, Cheng, X. | | Deposit date: | 1998-08-05 | | Release date: | 1998-11-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Structures of HhaI methyltransferase complexed with substrates containing mismatches at the target base.
Nat.Struct.Biol., 5, 1998
|
|
7MDZ
 
 | | 80S rabbit ribosome stalled with benzamide-CHX | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S21, ... | | Authors: | Koga, Y, Hoang, E.M, Park, Y, Keszei, A.F.A, Murray, J, Shao, S, Liau, B.B. | | Deposit date: | 2021-04-06 | | Release date: | 2021-09-01 | | Last modified: | 2021-09-15 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Discovery of C13-Aminobenzoyl Cycloheximide Derivatives that Potently Inhibit Translation Elongation.
J.Am.Chem.Soc., 143, 2021
|
|
7MQA
 
 | | Cryo-EM structure of the human SSU processome, state post-A1 | | Descriptor: | 18S rRNA, 40S ribosomal protein S11, 40S ribosomal protein S12, ... | | Authors: | Vanden Broeck, A, Singh, S, Klinge, S. | | Deposit date: | 2021-05-05 | | Release date: | 2021-09-22 | | Last modified: | 2021-09-29 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Nucleolar maturation of the human small subunit processome.
Science, 373, 2021
|
|
7MQ8
 
 | | Cryo-EM structure of the human SSU processome, state pre-A1 | | Descriptor: | 18S rRNA, 40S ribosomal protein S11, 40S ribosomal protein S12, ... | | Authors: | Vanden Broeck, A, Singh, S, Klinge, S. | | Deposit date: | 2021-05-05 | | Release date: | 2021-09-22 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Nucleolar maturation of the human small subunit processome.
Science, 373, 2021
|
|
7MQ9
 
 | | Cryo-EM structure of the human SSU processome, state pre-A1* | | Descriptor: | 18S rRNA, 40S ribosomal protein S11, 40S ribosomal protein S12, ... | | Authors: | Vanden Broeck, A, Singh, S, Klinge, S. | | Deposit date: | 2021-05-05 | | Release date: | 2021-09-22 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.87 Å) | | Cite: | Nucleolar maturation of the human small subunit processome.
Science, 373, 2021
|
|
7MJ1
 
 | | LarB, a carboxylase/hydrolase involved in synthesis of the cofactor for lactate racemase, in complex with NAD | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, MAGNESIUM ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Chatterjee, S, Rankin, J.A, Lagishetty, S, Hu, J, Hausinger, R.P. | | Deposit date: | 2021-04-19 | | Release date: | 2021-09-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.402 Å) | | Cite: | The LarB carboxylase/hydrolase forms a transient cysteinyl-pyridine intermediate during nickel-pincer nucleotide cofactor biosynthesis.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7MJ2
 
 | | LarB, a carboxylase/hydrolase involved in synthesis of the cofactor for lactate racemase, in complex with Zn | | Descriptor: | MAGNESIUM ION, Pyridinium-3,5-biscarboxylic acid mononucleotide synthase, ZINC ION | | Authors: | Chatterjee, S, Rankin, J.A, Lagishetty, S, Hu, J, Hausinger, R.P. | | Deposit date: | 2021-04-19 | | Release date: | 2021-09-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The LarB carboxylase/hydrolase forms a transient cysteinyl-pyridine intermediate during nickel-pincer nucleotide cofactor biosynthesis.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7MJ0
 
 | | LarB, a carboxylase/hydrolase involved in synthesis of the cofactor for lactate racemase, in complex with adenosine monophosphate AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, MAGNESIUM ION, Pyridinium-3,5-biscarboxylic acid mononucleotide synthase | | Authors: | Chatterjee, S, Rankin, J.A, Lagishetty, S, Hu, J, Hausinger, R.P. | | Deposit date: | 2021-04-19 | | Release date: | 2021-09-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | The LarB carboxylase/hydrolase forms a transient cysteinyl-pyridine intermediate during nickel-pincer nucleotide cofactor biosynthesis.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6Z6N
 
 | | Cryo-EM structure of human EBP1-80S ribosomes (focus on EBP1) | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S10, ... | | Authors: | Wells, J.N, Buschauer, R, Mackens-Kiani, T, Best, K, Kratzat, H, Berninghausen, O, Becker, T, Cheng, J, Beckmann, R. | | Deposit date: | 2020-05-28 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure and function of yeast Lso2 and human CCDC124 bound to hibernating ribosomes.
Plos Biol., 18, 2020
|
|
6Z6K
 
 | | Cryo-EM structure of yeast reconstituted Lso2 bound to 80S ribosomes | | Descriptor: | 18S rRNA, 25S rRNA, 40S ribosomal protein S0-A, ... | | Authors: | Wells, J.N, Buschauer, R, Mackens-Kiani, T, Best, K, Kratzat, H, Berninghausen, O, Becker, T, Cheng, J, Beckmann, R. | | Deposit date: | 2020-05-28 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure and function of yeast Lso2 and human CCDC124 bound to hibernating ribosomes.
Plos Biol., 18, 2020
|
|
6ZJE
 
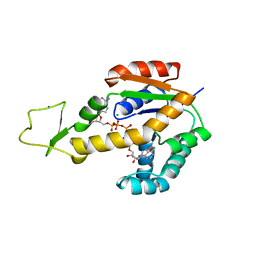 | | Crystal structure of human adenylate kinase 3, AK3, in complex with inhibitor Ap5A | | Descriptor: | BIS(ADENOSINE)-5'-PENTAPHOSPHATE, CHLORIDE ION, GTP:AMP phosphotransferase AK3, ... | | Authors: | Grundstrom, C, Rogne, P, Wolf-Watz, M, Sauer-Eriksson, A.E. | | Deposit date: | 2020-06-28 | | Release date: | 2020-09-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structural Basis for GTP versus ATP Selectivity in the NMP Kinase AK3.
Biochemistry, 59, 2020
|
|
6ZCE
 
 | | Structure of a yeast ABCE1-bound 43S pre-initiation complex | | Descriptor: | 18S ribosomal RNA (1719-MER), 40S ribosomal protein S0-A, 40S ribosomal protein S1-A, ... | | Authors: | Kratzat, H, Mackens-Kiani, T, Cheng, J, Berninghausen, O, Becker, T, Beckmann, R. | | Deposit date: | 2020-06-10 | | Release date: | 2020-10-07 | | Last modified: | 2021-01-13 | | Method: | ELECTRON MICROSCOPY (5.3 Å) | | Cite: | A structural inventory of native ribosomal ABCE1-43S pre-initiation complexes.
Embo J., 40, 2021
|
|
3SR0
 
 | |
3TF4
 
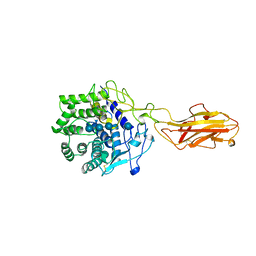 | | ENDO/EXOCELLULASE:CELLOTRIOSE FROM THERMOMONOSPORA | | Descriptor: | CALCIUM ION, T. FUSCA ENDO/EXO-CELLULASE E4 CATALYTIC DOMAIN AND CELLULOSE-BINDING DOMAIN, beta-D-glucopyranose, ... | | Authors: | Sakon, J, Wilson, D.B, Karplus, P.A. | | Deposit date: | 1997-05-30 | | Release date: | 1997-09-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and mechanism of endo/exocellulase E4 from Thermomonospora fusca.
Nat.Struct.Biol., 4, 1997
|
|
4ZGY
 
 | |
