3GMW
 
 | | Crystal Structure of Beta-Lactamse Inhibitory Protein-I (BLIP-I) in Complex with TEM-1 Beta-Lactamase | | Descriptor: | B-lactamase, Beta-lactamase inhibitory protein BLIP-I, PHOSPHATE ION | | Authors: | Lim, D.C, Gretes, M, Strynadka, N.C.J. | | Deposit date: | 2009-03-15 | | Release date: | 2009-03-31 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Insights into positive and negative requirements for protein-protein interactions by crystallographic analysis of the beta-lactamase inhibitory proteins BLIP, BLIP-I, and BLP.
J.Mol.Biol., 389, 2009
|
|
4NLW
 
 | | Poliovirus Polymerase - G289A/C290I Loop Mutant | | Descriptor: | ACETIC ACID, PENTAETHYLENE GLYCOL, RNA-directed RNA polymerase 3D-POL, ... | | Authors: | Sholders, A.J, Peersen, O.B. | | Deposit date: | 2013-11-14 | | Release date: | 2014-01-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Distinct conformations of a putative translocation element in poliovirus polymerase.
J.Mol.Biol., 426, 2014
|
|
4HIO
 
 | |
3GQ2
 
 | |
4NME
 
 | |
4HJ4
 
 | |
2OLK
 
 | | ABC Protein ArtP in complex with ADP-beta-S | | Descriptor: | 5'-O-[(R)-HYDROXY(THIOPHOSPHONOOXY)PHOSPHORYL]ADENOSINE, Amino acid ABC transporter | | Authors: | Thaben, P.F, Eckey, V, Scheffel, F, Saenger, W, Schneider, E, Vahedi-Faridi, A. | | Deposit date: | 2007-01-19 | | Release date: | 2008-01-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of the ATP-binding cassette (ABC) protein ArtP from Geobacillus stearothermophilus reveal a stable dimer in the post hydrolysis state and an asymmetry in the dimerization region
To be Published
|
|
3GUE
 
 | | Crystal Structure of UDP-glucose phosphorylase from Trypanosoma Brucei, (Tb10.389.0330) | | Descriptor: | (2R,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, GLYCEROL, SULFATE ION, ... | | Authors: | Wernimont, A.K, Marino, K, Lin, Y.H, Mackenzie, F, Kozieradzki, I, Cossar, D, Zhao, Y, Schapira, M, Bochkarev, A, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, Ferguson, M.A.J, Hui, R, Amani, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-03-29 | | Release date: | 2009-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal Structure of UDP-glucose phosphorylase from Trypanosoma Brucei, (Tb10.389.0330)
To be Published
|
|
5RSB
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000001674697 | | Descriptor: | 7-methyl-7H-pyrrolo[2,3-d]pyrimidin-4-amine, Non-structural protein 3 | | Authors: | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-09-28 | | Release date: | 2020-12-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
2EXU
 
 | | Crystal Structure of Saccharomyces cerevisiae transcription elongation factors Spt4-Spt5NGN domain | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ETHANOL, Transcription initiation protein SPT4/SPT5, ... | | Authors: | Xu, F, Guo, M, Fang, P, Teng, M, Niu, L. | | Deposit date: | 2005-11-08 | | Release date: | 2006-11-08 | | Last modified: | 2017-08-23 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Crystal Structure of Saccharomyces cerevisiae transcription elongation factors Spt4-Spt5NGN domain
To be published
|
|
4NR3
 
 | |
5RSQ
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000000158490 | | Descriptor: | 5-methyl-3-phenyl-1,2-oxazole-4-carboxylic acid, Non-structural protein 3 | | Authors: | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-09-28 | | Release date: | 2020-12-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
4NLY
 
 | | Poliovirus Polymerase - C290E Loop Mutant | | Descriptor: | ACETIC ACID, PENTAETHYLENE GLYCOL, RNA-directed RNA polymerase 3D-POL, ... | | Authors: | Sholders, A.J, Peersen, O.B. | | Deposit date: | 2013-11-14 | | Release date: | 2014-01-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Distinct conformations of a putative translocation element in poliovirus polymerase.
J.Mol.Biol., 426, 2014
|
|
5RT6
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000000156509 | | Descriptor: | 2-(3,4-dichlorophenyl)ethanoic acid, DIMETHYL SULFOXIDE, Non-structural protein 3 | | Authors: | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-09-28 | | Release date: | 2020-12-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
2OMT
 
 | | Crystal structure of InlA G194S+S/hEC1 complex | | Descriptor: | CALCIUM ION, CHLORIDE ION, Epithelial-cadherin; E-Cad/CTF1, ... | | Authors: | Wollert, T, Heinz, D.W, Schubert, W.D. | | Deposit date: | 2007-01-23 | | Release date: | 2007-08-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Thermodynamically reengineering the listerial invasion complex InlA/E-cadherin.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
4NMF
 
 | | Crystal structure of proline utilization A (PutA) from Geobacter sulfurreducens PCA inactivated by N-propargylglycine and complexed with menadione bisulfite | | Descriptor: | (2R)-2-methyl-1,4-dioxo-1,2,3,4-tetrahydronaphthalene-2-sulfonic acid, (2S)-2-methyl-1,4-dioxo-1,2,3,4-tetrahydronaphthalene-2-sulfonic acid, 1,2-ETHANEDIOL, ... | | Authors: | Singh, H, Tanner, J.J. | | Deposit date: | 2013-11-14 | | Release date: | 2014-02-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structures of the PutA peripheral membrane flavoenzyme reveal a dynamic substrate-channeling tunnel and the quinone-binding site.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2FDS
 
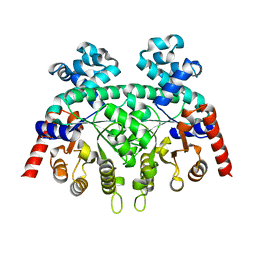 | | Crystal Structure of Plasmodium Berghei Orotidine 5'-monophosphate Decarboxylase (ortholog of Plasmodium falciparum PF10_0225) | | Descriptor: | IODIDE ION, orotidine-monophosphate-decarboxylase | | Authors: | Qiu, W, Dong, A, Wasney, G, Vedadi, M, Lew, J, Kozieradski, I, Alam, Z, Melone, M, Weigelt, J, Sundstrom, M, Edwards, A, Arrowsmith, C, Hui, R, Gao, M, Bochkarev, A, Artz, J.D, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-12-14 | | Release date: | 2005-12-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Genome-scale protein expression and structural biology of Plasmodium falciparum and related Apicomplexan organisms.
Mol.Biochem.Parasitol., 151, 2007
|
|
4HM5
 
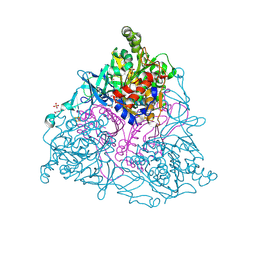 | | Naphthalene 1,2-Dioxygenase bound to indene | | Descriptor: | 1,2-ETHANEDIOL, 2,3-dihydro-1H-indene, FE (III) ION, ... | | Authors: | Ferraro, D.J, Ramaswamy, S. | | Deposit date: | 2012-10-17 | | Release date: | 2013-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Naphthalene 1,2-Dioxygenase bound to indene
To be Published
|
|
5RTM
 
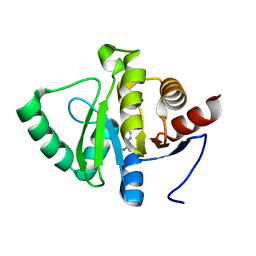 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000000002005 | | Descriptor: | Non-structural protein 3, PYRAZINE-2-CARBOXAMIDE | | Authors: | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-09-28 | | Release date: | 2020-12-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
3GV9
 
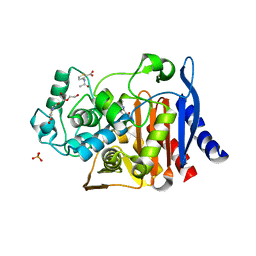 | |
3Q9Z
 
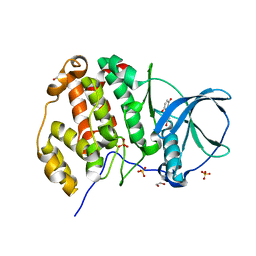 | | Crystal structure of human CK2 alpha in complex with Quinalizarin at pH 6.5 | | Descriptor: | 1,2,5,8-tetrahydroxyanthracene-9,10-dione, 1,2-ETHANEDIOL, Casein kinase II subunit alpha, ... | | Authors: | Battistutta, R, Ranchio, A, Papinutto, E. | | Deposit date: | 2011-01-10 | | Release date: | 2012-01-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and functional analysis of the flexible regions of the catalytic alpha-subunit of protein kinase CK2
To be Published
|
|
4HSN
 
 | | Crystal structure of DAH7PS from Neisseria meningitidis | | Descriptor: | 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase, MANGANESE (II) ION, PHOSPHOENOLPYRUVATE, ... | | Authors: | Cross, P.J, Pietersma, A.L, Allison, T.M, Wilson-Coutts, S.M, Cochrane, F.C, Parker, E.J. | | Deposit date: | 2012-10-30 | | Release date: | 2013-09-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Neisseria meningitidis expresses a single 3-deoxy-d-arabino-heptulosonate 7-phosphate synthase that is inhibited primarily by phenylalanine.
Protein Sci., 22, 2013
|
|
5RU0
 
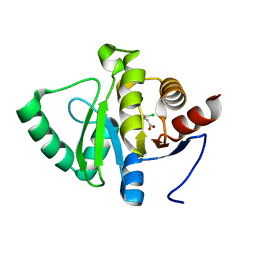 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000000388514 | | Descriptor: | (2,6-DICHLOROPHENYL)ACETIC ACID, Non-structural protein 3 | | Authors: | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-09-28 | | Release date: | 2020-12-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5RUH
 
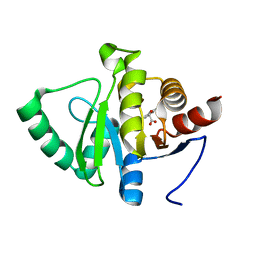 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000000123600 | | Descriptor: | (2,6-dichlorophenoxy)acetic acid, Non-structural protein 3 | | Authors: | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-09-28 | | Release date: | 2020-12-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5RUY
 
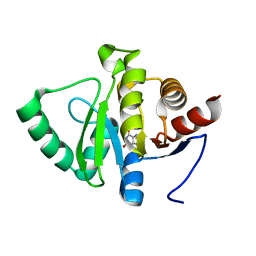 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000013517187 | | Descriptor: | Non-structural protein 3, XANTHINE | | Authors: | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-09-28 | | Release date: | 2020-12-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
